Experimental evolution
| Part of a series on |
| Evolutionary biology |
|---|
 |
Experimental evolution is the use of laboratory experiments or controlled field manipulations to explore evolutionary dynamics.[1] Evolution may be observed in the laboratory as individuals/populations adapt to new environmental conditions by natural selection.
There are two different ways in which adaptation can arise in experimental evolution. One is via an individual organism gaining a novel beneficial mutation.[2] The other is from allele frequency change in standing genetic variation already present in a population of organisms.[2] Other evolutionary forces outside of mutation and natural selection can also play a role or be incorporated into experimental evolution studies, such as genetic drift and gene flow.[3]
The organism used is decided by the experimenter, based on the hypothesis to be tested. Many generations are required for adaptive mutation to occur, and experimental evolution via mutation is carried out in viruses or unicellular organisms with rapid generation times, such as bacteria and asexual clonal yeast.[1][4][5] Polymorphic populations of asexual or sexual yeast,[2] and multicellular eukaryotes like Drosophila, can adapt to new environments through allele frequency change in standing genetic variation.[3] Organisms with longer generations times, although costly, can be used in experimental evolution. Laboratory studies with foxes[6] and with rodents (see below) have shown that notable adaptations can occur within as few as 10–20 generations and experiments with wild guppies have observed adaptations within comparable numbers of generations.[7]
More recently, experimentally evolved individuals or populations are often analyzed using whole genome sequencing,[8][9] an approach known as Evolve and Resequence (E&R).[10] E&R can identify mutations that lead to adaptation in clonal individuals or identify alleles that changed in frequency in polymorphic populations, by comparing the sequences of individuals/populations before and after adaptation.[2] The sequence data makes it possible to pinpoint the site in a DNA sequence that a mutation/allele frequency change occurred to bring about adaptation.[10][9][2] The nature of the adaptation and functional follow up studies can shed insight into what effect the mutation/allele has on phenotype.
History
[edit]Domestication and breeding
[edit]
Unwittingly, humans have carried out evolution experiments for as long as they have been domesticating plants and animals. Selective breeding of plants and animals has led to varieties that differ dramatically from their original wild-type ancestors. Examples are the cabbage varieties, maize, or the large number of different dog breeds. The power of human breeding to create varieties with extreme differences from a single species was already recognized by Charles Darwin. In fact, he started out his book The Origin of Species with a chapter on variation in domestic animals. In this chapter, Darwin discussed in particular the pigeon.
Altogether at least a score of pigeons might be chosen, which if shown to an ornithologist, and he were told that they were wild birds, would certainly, I think, be ranked by him as well-defined species. Moreover, I do not believe that any ornithologist would place the English carrier, the short-faced tumbler, the runt, the barb, pouter, and fantail in the same genus; more especially as in each of these breeds several truly-inherited sub-breeds, or species as he might have called them, could be shown him. (...) I am fully convinced that the common opinion of naturalists is correct, namely, that all have descended from the rock-pigeon (Columba livia), including under this term several geographical races or sub-species, which differ from each other in the most trifling respects.
— Charles Darwin, The Origin of Species
Early
[edit]
One of the first to carry out a controlled evolution experiment was William Dallinger. In the late 19th century, he cultivated small unicellular organisms in a custom-built incubator over a time period of seven years (1880–1886). Dallinger slowly increased the temperature of the incubator from an initial 60 °F up to 158 °F. The early cultures had shown clear signs of distress at a temperature of 73 °F, and were certainly not capable of surviving at 158 °F. The organisms Dallinger had in his incubator at the end of the experiment, on the other hand, were perfectly fine at 158 °F. However, these organisms would no longer grow at the initial 60 °F. Dallinger concluded that he had found evidence for Darwinian adaptation in his incubator, and that the organisms had adapted to live in a high-temperature environment. Dallinger's incubator was accidentally destroyed in 1886, and Dallinger could not continue this line of research.[11][12]
From the 1880s to 1980, experimental evolution was intermittently practiced by a variety of evolutionary biologists, including the highly influential Theodosius Dobzhansky. Like other experimental research in evolutionary biology during this period, much of this work lacked extensive replication and was carried out only for relatively short periods of evolutionary time.[13]
Modern
[edit]Experimental evolution has been used in various formats to understand underlying evolutionary processes in a controlled system. Experimental evolution has been performed on multicellular[14] and unicellular[15] eukaryotes, prokaryotes,[16] and viruses.[17] Similar works have also been performed by directed evolution of individual enzyme,[18][19] ribozyme[20] and replicator[21][22] genes.
Aphids
[edit]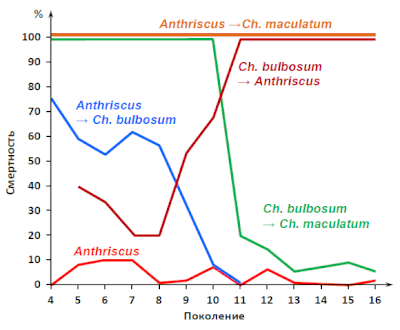
In the 1950s, the Soviet biologist Georgy Shaposhnikov conducted experiments on aphids of the Dysaphis genus. By transferring them to plants normally nearly or completely unsuitable for them, he had forced populations of parthenogenetic descendants to adapt to the new food source to the point of reproductive isolation from the regular populations of the same species.[23]
Fruit flies
[edit]One of the first of a new wave of experiments using this strategy was the laboratory "evolutionary radiation" of Drosophila melanogaster populations that Michael R. Rose started in February, 1980.[24] This system started with ten populations, five cultured at later ages, and five cultured at early ages. Since then more than 200 different populations have been created in this laboratory radiation, with selection targeting multiple characters. Some of these highly differentiated populations have also been selected "backward" or "in reverse," by returning experimental populations to their ancestral culture regime. Hundreds of people have worked with these populations over the better part of three decades. Much of this work is summarized in the papers collected in the book Methuselah Flies.[25]
The early experiments in flies were limited to studying phenotypes but the molecular mechanisms, i.e., changes in DNA that facilitated such changes, could not be identified. This changed with genomics technology.[26] Subsequently, Thomas Turner coined the term Evolve and Resequence (E&R)[10] and several studies used E&R approach with mixed success.[27][28] One of the more interesting experimental evolution studies was conducted by Gabriel Haddad's group at UC San Diego, where Haddad and colleagues evolved flies to adapt to low oxygen environments, also known as hypoxia.[29] After 200 generations, they used E&R approach to identify genomic regions that were selected by natural selection in the hypoxia adapted flies.[30] More recent experiments are following up E&R predictions with RNAseq[31] and genetic crosses.[9] Such efforts in combining E&R with experimental validations should be powerful in identifying genes that regulate adaptation in flies.
Much recently the experimental evolution in flies have taken the course to address the molecular mechanisms[32][33] and in doing so it might pave way to understand physiology of an organism better and thus redefine disease therapeutics.[34]
Microbes
[edit]Many microbial species have short generation times, easily sequenced genomes, and well-understood biology. They are therefore commonly used for experimental evolution studies. The bacterial species most commonly used for experimental evolution include P. fluorescens,[35] Pseudomonas aeruginosa,[36] Enterococcus faecalis [37] and E. coli (see below), while the Yeast S. cerevisiae has been used as a model for the study of eukaryotic evolution.[38]
Lenski's E. coli experiment
[edit]One of the most widely known examples of laboratory bacterial evolution is the long-term E.coli experiment of Richard Lenski. On February 24, 1988, Lenski started growing twelve lineages of E. coli under identical growth conditions.[39][40] When one of the populations evolved the ability to aerobically metabolize citrate from the growth medium and showed greatly increased growth,[41] this provided a dramatic observation of evolution in action. The experiment continues to this day, and is now the longest-running (in terms of generations) controlled evolution experiment ever undertaken.[citation needed] Since the inception of the experiment, the bacteria have grown for more than 60,000 generations. Lenski and colleagues regularly publish updates on the status of the experiments.[42]
Leishmania donovani
[edit]Bussotti and collaborators isolated amastigotes from Leishmania donovani and cultured them in vitro for 3800 generations (36 weeks). The culture of these parasites showed how they adapted to in vitro conditions by compensating for the loss of a NIMA-related kinase, important for the correct progression of mitosis, by increasing the expression of another orthologous kinase as the culture generations progressed. Furthermore, it was observed how L. donovani has been adapted to in vitro culture by reducing the expression of 23 transcripts related to flagellar biogenesis and increasing the expression of ribosomal protein clusters and non-coding RNAs such as nucleolar small RNAs. Flagella are considered less necessary by the parasite in in vitro culture and therefore the progression of generations leads to their elimination, causing an energy saving due to lower motility so that proliferation and growth rate in culture is higher. The amplified snoRNAs also lead to increased ribosomal biosynthesis, increased protein biosynthesis and thus increased growth rate of the culture. These adaptations observed over generations of parasites are governed by copy number variations (CNV) and epistatic interactions between affected genes, and allow us to justify Leishmania genomic instability through its post-transcriptional regulation of gene expression.[43]
Laboratory house mice
[edit]
In 1993, Theodore Garland, Jr. and colleagues started a long-term experiment that involves selective breeding of mice for high voluntary activity levels on running wheels.[44] This experiment also continues to this day (> 105 generations). Mice from the four replicate "High Runner" lines evolved to run almost three times as many running-wheel revolutions per day compared with the four unselected control lines of mice, mainly by running faster than the control mice rather than running for more minutes/day. However, the High Runner lines have evolved in somewhat different ways, with some emphasizing running speed versus duration or vice versa, thus demonstrating "multiple solutions" [45] that seem to be based partly in evolved muscle characteristics.[46]

The HR mice have an elevated endurance running ability [47] and maximal aerobic capacity [48] when tested on a motorized treadmill. They also exhibit alterations in motivation and the reward system of the brain. Pharmacological studies point to alterations in dopamine function and the endocannabinoid system.[49] The High Runner lines have been proposed as a model to study human attention-deficit hyperactivity disorder (ADHD), and administration of Ritalin reduces their wheel running approximately to the levels of control mice.[50]
Multidirectional selection on bank voles
[edit]In 2005 Paweł Koteja with Edyta Sadowska and colleagues from the Jagiellonian University (Poland) started a multidirectional selection on a non-laboratory rodent, the bank vole Myodes (= Clethrionomys) glareolus.[51] The voles are selected for three distinct traits, which played important roles in the adaptive radiation of terrestrial vertebrates: high maximum rate of aerobic metabolism, predatory propensity, and herbivorous capability. Aerobic lines are selected for the maximum rate of oxygen consumption achieved during swimming at 38°C; Predatory lines – for a short time to catch live crickets; Herbivorous lines – for capability to maintain body mass when fed a low-quality diet “diluted” with dried, powdered grass. Four replicate lines are maintained for each of the three selection directions and another four as unselected Controls.
After approximately 20 generations of selective breeding, voles from the Aerobic lines evolved a 60% higher swim-induced metabolic rate than voles from the unselected Control lines. Although the selection protocol does not impose a thermoregulatory burden, both the basal metabolic rate and thermogenic capacity increased in the Aerobic lines.[52][53] Thus, the results have provided some support for the “aerobic capacity model” for the evolution of endothermy in mammals.
More than 85% of the Predatory voles capture the crickets, compared to only about 15% of unselected Control voles, and they catch the crickets faster. The increased predatory behavior is associated with a more proactive coping style (“personality”).[54]
During the test with low-quality diet, the Herbivorous voles lose approximately 2 grams less mass (approximately 10% of the original body mass) than the Control ones. The Herbivorous voles have an altered composition of the bacterial microbiome in their caecum.[55] Thus, the selection has resulted in evolution of the entire holobiome, and the experiment may offer a laboratory model of hologenome evolution.
Synthetic biology
[edit]Synthetic biology offers unique opportunities for experimental evolution, facilitating the interpretation of evolutionary changes by inserting genetic modules into host genomes and applying selection specifically targeting such modules. Synthetic biological circuits inserted into the genome of Escherichia coli[56] or the budding yeast Saccharomyces cerevisiae[57] degrade (lose function) during laboratory evolution. With appropriate selection, mechanisms underlying the evolutionary regain of lost biological function can be studied.[58] Experimental evolution of mammalian cells harboring synthetic gene circuits[59] reveals the role of cellular heterogeneity in the evolution of drug resistance, with implications for chemotherapy resistance of cancer cells.
Other examples
[edit]Stickleback fish have both marine and freshwater species, the freshwater species evolving since the last ice age. Freshwater species can survive colder temperatures. Scientists tested to see if they could reproduce this evolution of cold-tolerance by keeping marine sticklebacks in cold freshwater. It took the marine sticklebacks only three generations to evolve to match the 2.5 degree Celsius improvement in cold-tolerance found in wild freshwater sticklebacks.[60]
Microbial cells [61] and recently mammalian cells [62] are evolved under nutrient limiting conditions to study their metabolic response and engineer cells for useful characteristics.
For teaching
[edit]Because of their rapid generation times microbes offer an opportunity to study microevolution in the classroom. A number of exercises involving bacteria and yeast teach concepts ranging from the evolution of resistance[63] to the evolution of multicellularity.[64] With the advent of next-generation sequencing technology it has become possible for students to conduct an evolutionary experiment, sequence the evolved genomes, and to analyze and interpret the results.[65]
See also
[edit]References
[edit]- ^ a b "Experimental Evolution". Nature.
- ^ a b c d e Long A, Liti G, Luptak A, Tenaillon O (October 2015). "Elucidating the molecular architecture of adaptation via evolve and resequence experiments". Nature Reviews. Genetics. 16 (10): 567–582. doi:10.1038/nrg3937. PMC 4733663. PMID 26347030.
- ^ a b Kawecki, Tadeusz J.; Lenski, Richard E.; Ebert, Dieter; Hollis, Brian; Olivieri, Isabelle; Whitlock, Michael C. (October 2012). "Experimental evolution" (PDF). Trends in Ecology & Evolution. 27 (10): 547–560. Bibcode:2012TEcoE..27..547K. doi:10.1016/j.tree.2012.06.001. PMID 22819306.
- ^ Buckling A, Craig Maclean R, Brockhurst MA, Colegrave N (February 2009). "The Beagle in a bottle". Nature. 457 (7231): 824–829. Bibcode:2009Natur.457..824B. doi:10.1038/nature07892. PMID 19212400. S2CID 205216404.
- ^ Elena SF, Lenski RE (June 2003). "Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation". Nature Reviews. Genetics. 4 (6): 457–469. doi:10.1038/nrg1088. PMID 12776215. S2CID 209727.
- ^ Trut, Lyudmila (1999). "Early Canid Domestication: The Farm-Fox Experiment". American Scientist. 87 (2): 160–169. doi:10.1511/1999.2.160. JSTOR 27857815.
- ^ Reznick DN, Shaw FH, Rodd FH, Shaw RG (March 1997). "Evaluation of the Rate of Evolution in Natural Populations of Guppies (Poecilia reticulata)". Science. 275 (5308): 1934–1937. doi:10.1126/science.275.5308.1934. PMID 9072971. S2CID 18480502.
- ^ Barrick JE, Lenski RE (December 2013). "Genome dynamics during experimental evolution". Nature Reviews. Genetics. 14 (12): 827–839. doi:10.1038/nrg3564. PMC 4239992. PMID 24166031.
- ^ a b c Jha AR, Miles CM, Lippert NR, Brown CD, White KP, Kreitman M (October 2015). "Whole-Genome Resequencing of Experimental Populations Reveals Polygenic Basis of Egg-Size Variation in Drosophila melanogaster". Molecular Biology and Evolution. 32 (10): 2616–2632. doi:10.1093/molbev/msv136. PMC 4576704. PMID 26044351.
- ^ a b c Turner TL, Stewart AD, Fields AT, Rice WR, Tarone AM (March 2011). "Population-based resequencing of experimentally evolved populations reveals the genetic basis of body size variation in Drosophila melanogaster". PLOS Genetics. 7 (3): e1001336. doi:10.1371/journal.pgen.1001336. PMC 3060078. PMID 21437274.
- ^ Haas JW (January 2000). "The Reverend Dr William Henry Dallinger, F.R.S. (1839-1909)". Notes and Records of the Royal Society of London. 54 (1): 53–65. doi:10.1098/rsnr.2000.0096. PMID 11624308. S2CID 145758182.
- ^ Zimmer C (2011). "Darwin Under the Microscope: Witnessing Evolution in Microbes" (PDF). In Losos J (ed.). In the Light of Evolution: Essays from the Laboratory and Field. W. H. Freeman. pp. 42–43. ISBN 978-0-9815194-9-4.
- ^ Dobzhansky T, Pavlovsky O (1957). "An experimental study of interaction between genetic drift and natural selection". Evolution. 11 (3): 311–319. doi:10.2307/2405795. JSTOR 2405795.
- ^ Marden JH, Wolf MR, Weber KE (November 1997). "Aerial performance of Drosophila melanogaster from populations selected for upwind flight ability". The Journal of Experimental Biology. 200 (Pt 21): 2747–2755. doi:10.1242/jeb.200.21.2747. PMID 9418031.
- ^ Ratcliff WC, Denison RF, Borrello M, Travisano M (January 2012). "Experimental evolution of multicellularity". Proceedings of the National Academy of Sciences of the United States of America. 109 (5): 1595–1600. Bibcode:2012PNAS..109.1595R. doi:10.1073/pnas.1115323109. PMC 3277146. PMID 22307617.
- ^ Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, et al. (October 2009). "Genome evolution and adaptation in a long-term experiment with Escherichia coli". Nature. 461 (7268): 1243–1247. Bibcode:2009Natur.461.1243B. doi:10.1038/nature08480. PMID 19838166. S2CID 4330305.
- ^ Heineman RH, Molineux IJ, Bull JJ (August 2005). "Evolutionary robustness of an optimal phenotype: re-evolution of lysis in a bacteriophage deleted for its lysin gene". Journal of Molecular Evolution. 61 (2): 181–191. Bibcode:2005JMolE..61..181H. doi:10.1007/s00239-004-0304-4. PMID 16096681. S2CID 31230414.
- ^ Bloom JD, Arnold FH (June 2009). "In the light of directed evolution: pathways of adaptive protein evolution". Proceedings of the National Academy of Sciences of the United States of America. 106 (Suppl 1): 9995–10000. doi:10.1073/pnas.0901522106. PMC 2702793. PMID 19528653.
- ^ Moses AM, Davidson AR (May 2011). "In vitro evolution goes deep". Proceedings of the National Academy of Sciences of the United States of America. 108 (20): 8071–8072. Bibcode:2011PNAS..108.8071M. doi:10.1073/pnas.1104843108. PMC 3100951. PMID 21551096.
- ^ Salehi-Ashtiani K, Szostak JW (November 2001). "In vitro evolution suggests multiple origins for the hammerhead ribozyme". Nature. 414 (6859): 82–84. Bibcode:2001Natur.414...82S. doi:10.1038/35102081. PMID 11689947. S2CID 4401483.
- ^ Sumper M, Luce R (January 1975). "Evidence for de novo production of self-replicating and environmentally adapted RNA structures by bacteriophage Qbeta replicase". Proceedings of the National Academy of Sciences of the United States of America. 72 (1): 162–166. Bibcode:1975PNAS...72..162S. doi:10.1073/pnas.72.1.162. PMC 432262. PMID 1054493.
- ^ Mills DR, Peterson RL, Spiegelman S (July 1967). "An extracellular Darwinian experiment with a self-duplicating nucleic acid molecule". Proceedings of the National Academy of Sciences of the United States of America. 58 (1): 217–224. Bibcode:1967PNAS...58..217M. doi:10.1073/pnas.58.1.217. PMC 335620. PMID 5231602.
- ^ Shaposhnikov GK (1966). "Origin and breakdown of reproductive isolation and the criterion of the species" (PDF). Entomological Review. 45: 1–8. Archived from the original (PDF) on 2013-09-08.
- ^ Rose MR (May 1984). "Artificial Selection on a Fitness-Component in Drosophila Melanogaster". Evolution; International Journal of Organic Evolution. 38 (3): 516–526. doi:10.2307/2408701. JSTOR 2408701. PMID 28555975.
- ^ Rose MR, Passananti HB, Matos M (2004). Methuselah Flies. Singapore: World Scientific. doi:10.1142/5457. ISBN 978-981-238-741-7.
- ^ Burke MK, Dunham JP, Shahrestani P, Thornton KR, Rose MR, Long AD (September 2010). "Genome-wide analysis of a long-term evolution experiment with Drosophila". Nature. 467 (7315): 587–590. Bibcode:2010Natur.467..587B. doi:10.1038/nature09352. PMID 20844486. S2CID 205222217.
- ^ Schlötterer C, Tobler R, Kofler R, Nolte V (November 2014). "Sequencing pools of individuals - mining genome-wide polymorphism data without big funding". Nature Reviews. Genetics. 15 (11): 749–763. doi:10.1038/nrg3803. PMID 25246196. S2CID 35827109.
- ^ Schlötterer C, Kofler R, Versace E, Tobler R, Franssen SU (May 2015). "Combining experimental evolution with next-generation sequencing: a powerful tool to study adaptation from standing genetic variation". Heredity. 114 (5): 431–440. doi:10.1038/hdy.2014.86. PMC 4815507. PMID 25269380.
- ^ Zhou D, Xue J, Chen J, Morcillo P, Lambert JD, White KP, Haddad GG (May 2007). "Experimental selection for Drosophila survival in extremely low O(2) environment". PLOS ONE. 2 (5): e490. Bibcode:2007PLoSO...2..490Z. doi:10.1371/journal.pone.0000490. PMC 1871610. PMID 17534440.
- ^ Zhou D, Udpa N, Gersten M, Visk DW, Bashir A, Xue J, et al. (February 2011). "Experimental selection of hypoxia-tolerant Drosophila melanogaster". Proceedings of the National Academy of Sciences of the United States of America. 108 (6): 2349–2354. Bibcode:2011PNAS..108.2349Z. doi:10.1073/pnas.1010643108. PMC 3038716. PMID 21262834.
- ^ Remolina SC, Chang PL, Leips J, Nuzhdin SV, Hughes KA (November 2012). "Genomic basis of aging and life-history evolution in Drosophila melanogaster". Evolution; International Journal of Organic Evolution. 66 (11): 3390–3403. doi:10.1111/j.1558-5646.2012.01710.x. PMC 4539122. PMID 23106705.
- ^ Shrivastava, Nidhi Krishna; Shakarad, Mallikarjun N. (May 2023). "Correlated responses in basal immune function in response to selection for fast development in Drosophila melanogaster". Journal of Evolutionary Biology. 36 (5): 816–828. doi:10.1111/jeb.14176. PMID 37073855.
- ^ Shrivastava, Nidhi Krishna; Chauhan, Namita; Shakarad, Mallikarjun N. (December 2022). "Heightened immune surveillance in Drosophila melanogaster populations selected for faster development and extended longevity". Heliyon. 8 (12): e12090. Bibcode:2022Heliy...812090S. doi:10.1016/j.heliyon.2022.e12090. PMC 9761728. PMID 36544838.
- ^ Shrivastava, Nidhi Krishna; Farand, Abhishek Kumar; Shakarad, Mallikarjun N. (December 2022). "Long-term selection for faster development and early reproduction leads to up-regulation of genes involved in redox homeostasis". Advances in Redox Research. 6: 100045. doi:10.1016/j.arres.2022.100045.
- ^ Rainey PB, Travisano M (July 1998). "Adaptive radiation in a heterogeneous environment". Nature. 394 (6688): 69–72. Bibcode:1998Natur.394...69R. doi:10.1038/27900. PMID 9665128. S2CID 40896184.
- ^ Chua SL, Ding Y, Liu Y, Cai Z, Zhou J, Swarup S, et al. (November 2016). "Reactive oxygen species drive evolution of pro-biofilm variants in pathogens by modulating cyclic-di-GMP levels". Open Biology. 6 (11): 160162. doi:10.1098/rsob.160162. PMC 5133437. PMID 27881736.
- ^ Ma Y, Chua SL (2021-11-15). "No collateral antibiotic sensitivity by alternating antibiotic pairs". The Lancet Microbe. 3 (1): e7. doi:10.1016/S2666-5247(21)00270-6. ISSN 2666-5247. PMID 35544116. S2CID 244147577.
- ^ Rainey PB, Travisano M (July 1998). "Adaptive radiation in a heterogeneous environment". Nature. 394 (6688): 69–72. Bibcode:2013Natur.500..571L. doi:10.1038/nature12344. PMC 3758440. PMID 9665128.
- ^ Lenski RE, Rose MR, Simpson SC, Tadler SC (1991-12-01). "Long-Term Experimental Evolution in Escherichia coli. I. Adaptation and Divergence During 2,000 Generations". The American Naturalist. 138 (6): 1315–1341. doi:10.1086/285289. ISSN 0003-0147. S2CID 83996233.
- ^ Fox JW, Lenski RE (June 2015). "From Here to Eternity--The Theory and Practice of a Really Long Experiment". PLOS Biology. 13 (6): e1002185. doi:10.1371/journal.pbio.1002185. PMC 4477892. PMID 26102073.
- ^ Blount ZD, Borland CZ, Lenski RE (June 2008). "Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli". Proceedings of the National Academy of Sciences of the United States of America. 105 (23): 7899–7906. Bibcode:2008PNAS..105.7899B. doi:10.1073/pnas.0803151105. PMC 2430337. PMID 18524956.
- ^ Lenski RE. "E. coli Long-term Experimental Evolution Project Site". Michigan State University. Archived from the original on 2017-07-27. Retrieved 2004-07-08.
- ^ Bussotti, Giovanni; Piel, Laura; Pescher, Pascale; Domagalska, Malgorzata A.; Rajan, K. Shanmugha; Cohen-Chalamish, Smadar; Doniger, Tirza; Hiregange, Disha-Gajanan; Myler, Peter J.; Unger, Ron; Michaeli, Shulamit; Späth, Gerald F. (21 December 2021). "Genome instability drives epistatic adaptation in the human pathogen Leishmania". Proceedings of the National Academy of Sciences. 118 (51): e2113744118. Bibcode:2021PNAS..11813744B. doi:10.1073/pnas.2113744118. ISSN 0027-8424. PMC 8713814. PMID 34903666.
- ^ Swallow, John G.; Carter, Patrick A.; Garland, Jr., Theodore (1998). "Artificial Selection for Increased Wheel-Running Behavior in House Mice". Behavior Genetics. 28 (3): 227–237. doi:10.1023/a:1021479331779. PMID 9670598.
- ^ Garland, Jr., T.; Kelly, S. A.; Malisch, J. L.; Kolb, E. M.; Hannon, R. M.; Keeney, B. K.; Van Cleave, S. L.; Middleton, K. M. (2011). "How to run far: multiple solutions and sex-specific responses to selective breeding for high voluntary activity levels". Proceedings of the Royal Society B: Biological Sciences. 278 (1705): 574–581. doi:10.1098/rspb.2010.1584. PMC 3025687. PMID 20810439.
- ^ Castro, A. A.; Garland, Jr., T.; Ahmed, S.; Holt, N. C. (2022). "Trade-offs in muscle physiology in selectively bred High Runner mice". Journal of Experimental Biology. 225 (23): jeb244083. doi:10.1242/jeb.244083. PMC 9789404. PMID 36408738.
- ^ Meek, T. E.; Lonquich, B. P.; Hannon, R. M.; Garland, Jr., T. (2009). "Endurance capacity of mice selectively bred for high voluntary wheel running". Journal of Experimental Biology. 212 (18): 2908–2917. doi:10.1242/jeb.028886. PMID 19717672.
- ^ Schwartz, N. E.; McNamara, M. P.; Orozco, J. M.; Rashid, J. O.; Thai, A. P.; Garland, Jr., T. (2023). "Selective breeding for high voluntary exercise in mice increases maximal (V̇O2,max) but not basal metabolic rate". Journal of Experimental Biology. 226 (15): jeb245256. doi:10.1242/jeb.245256. PMID 37439323.
- ^ Keeney BK, Raichlen DA, Meek TH, Wijeratne RS, Middleton KM, Gerdeman GL, Garland T (December 2008). "Differential response to a selective cannabinoid receptor antagonist (SR141716: rimonabant) in female mice from lines selectively bred for high voluntary wheel-running behaviour". Behavioural Pharmacology. 19 (8): 812–820. doi:10.1097/FBP.0b013e32831c3b6b. PMID 19020416. S2CID 16215160.
- ^ Rhodes, J. S.; Garland, Jr., T. (2003). "Differential sensitivity to acute administration of Ritalin, apomorphine, SCH 23390, and raclopride in mice selectively bred for hyperactive wheel-running behavior". Psychopharmacology. 167 (3): 242–250. doi:10.1007/s00213-003-1399-9. PMID 12669177.
- ^ Sadowska ET, Baliga-Klimczyk K, Chrzaścik KM, Koteja P (2008). "Laboratory model of adaptive radiation: a selection experiment in the bank vole". Physiological and Biochemical Zoology. 81 (5): 627–640. doi:10.1086/590164. PMID 18781839. S2CID 20125314.
- ^ Sadowska ET, Stawski C, Rudolf A, Dheyongera G, Chrząścik KM, Baliga-Klimczyk K, Koteja P (May 2015). "Evolution of basal metabolic rate in bank voles from a multidirectional selection experiment". Proceedings. Biological Sciences. 282 (1806): 20150025. doi:10.1098/rspb.2015.0025. PMC 4426621. PMID 25876844.
- ^ Dheyongera G, Grzebyk K, Rudolf AM, Sadowska ET, Koteja P (April 2016). "The effect of chlorpyrifos on thermogenic capacity of bank voles selected for increased aerobic exercise metabolism". Chemosphere. 149: 383–390. Bibcode:2016Chmsp.149..383D. doi:10.1016/j.chemosphere.2015.12.120. PMID 26878110.
- ^ Maiti U, Sadowska ET, ChrzĄścik KM, Koteja P (August 2019). "Experimental evolution of personality traits: open-field exploration in bank voles from a multidirectional selection experiment". Current Zoology. 65 (4): 375–384. doi:10.1093/cz/zoy068. PMC 6688576. PMID 31413710.
- ^ Kohl KD, Sadowska ET, Rudolf AM, Dearing MD, Koteja P (2016). "Experimental Evolution on a Wild Mammal Species Results in Modifications of Gut Microbial Communities". Frontiers in Microbiology. 7: 634. doi:10.3389/fmicb.2016.00634. PMC 4854874. PMID 27199960.
- ^ Sleight SC, Bartley BA, Lieviant JA, Sauro HM (November 2010). "Designing and engineering evolutionary robust genetic circuits". Journal of Biological Engineering. 4: 12. doi:10.1186/1754-1611-4-12. PMC 2991278. PMID 21040586.
- ^ González C, Ray JC, Manhart M, Adams RM, Nevozhay D, Morozov AV, Balázsi G (August 2015). "Stress-response balance drives the evolution of a network module and its host genome". Molecular Systems Biology. 11 (8): 827. doi:10.15252/msb.20156185. PMC 4562500. PMID 26324468.
- ^ Kheir Gouda M, Manhart M, Balázsi G (December 2019). "Evolutionary regain of lost gene circuit function". Proceedings of the National Academy of Sciences. 116 (50): 25162–25171. Bibcode:2019PNAS..11625162K. doi:10.1073/pnas.1912257116. PMC 6911209. PMID 31754027.
- ^ Farquhar KS, Charlebois DA, Szenk M, Cohen J, Nevozhay D, Balázsi G (June 2019). "Role of network-mediated stochasticity in mammalian drug resistance". Nature Communications. 10 (1): 2766. doi:10.1038/s41467-019-10330-w. PMC 6591227. PMID 31235692.
- ^ Barrett RD, Paccard A, Healy TM, Bergek S, Schulte PM, Schluter D, Rogers SM (January 2011). "Rapid evolution of cold tolerance in stickleback". Proceedings. Biological Sciences. 278 (1703): 233–238. doi:10.1098/rspb.2010.0923. PMC 3013383. PMID 20685715.
- ^ Dragosits M, Mattanovich D (July 2013). "Adaptive laboratory evolution -- principles and applications for biotechnology". Microbial Cell Factories. 12 (1): 64. doi:10.1186/1475-2859-12-64. PMC 3716822. PMID 23815749.
- ^ Hyman P (January 2014). "Bacteriophage as instructional organisms in introductory biology labs". Bacteriophage. 4 (1): e27336. doi:10.4161/bact.27336. PMC 3895413. PMID 24478938.
- ^ Ratcliff WC, Raney A, Westreich S, Cotner S (2014). "A Novel Laboratory Activity for Teaching about the Evolution of Multicellularity". The American Biology Teacher. 76 (2): 81–87. doi:10.1525/abt.2014.76.2.3. ISSN 0002-7685. S2CID 86079463.
- ^ Mikheyev, Alexander S; Arora, Jigyasa (9 September 2015). Using experimental evolution and next-generation sequencing to teach bench and bioinformatic skills (Preprint). doi:10.7287/peerj.preprints.1356v1.
Further reading
[edit]- Bennett AF (2003). "Experimental evolution and the Krogh principle: generating biological novelty for functional and genetic analyses". Physiological and Biochemical Zoology. 76 (1): 1–11. doi:10.1086/374275. PMID 12695982. S2CID 9032244.
- Dallinger WH (April 1887). "The president's address". Journal of the Royal Microscopical Society. 7 (2): 185–99. doi:10.1111/j.1365-2818.1887.tb01566.x.
- Garland Jr T (2003). "Selection experiments: an under-utilized tool in biomechanics and organismal biology." (PDF). In Bels VL, Gasc JP, Casinos A (eds.). Vertebrate biomechanics and evolution. Oxford, UK: BIOS Scientific Publishers. pp. 23–56. Archived from the original (PDF) on 2015-09-23. Retrieved 2007-02-10.
- Garland Jr T, Rose MR, eds. (2009). Experimental evolution: concepts, methods, and applications of selection experiments. Berkeley, California: University of California Press. ISBN 978-0-520-26180-8.
- Gibbs AG (October 1999). "Laboratory selection for the comparative physiologist". The Journal of Experimental Biology. 202 (Pt 20): 2709–2718. doi:10.1242/jeb.202.20.2709. PMID 10504307.
- Lenski, Richard E. (2003). "Phenotypic and Genomic Evolution during a 20,000-Generation Experiment with the Bacterium Escherichia coli". Plant Breeding Reviews. pp. 225–265. doi:10.1002/9780470650288.ch8. ISBN 978-0-471-46892-9.
- Lenski RE, Rose MR, Simpson SC, Tadler SC (1991). "Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations". American Naturalist. 138 (6): 1315–1341. doi:10.1086/285289. S2CID 83996233.
- McKenzie JA, Batterham P (May 1994). "The genetic, molecular and phenotypic consequences of selection for insecticide resistance". Trends in Ecology & Evolution. 9 (5): 166–169. Bibcode:1994TEcoE...9..166M. doi:10.1016/0169-5347(94)90079-5. PMID 21236810.
- Reznick DN, Bryant MJ, Roff D, Ghalambor CK, Ghalambor DE (October 2004). "Effect of extrinsic mortality on the evolution of senescence in guppies". Nature. 431 (7012): 1095–1099. Bibcode:2004Natur.431.1095R. doi:10.1038/nature02936. PMID 15510147. S2CID 205210169.
- Rose MR, Passananti HB, Matos M, eds. (2004). Methuselah flies: A case study in the evolution of aging. Singapore: World Scientific Publishing.
- Swallow JG, Garland T (June 2005). "Selection Experiments as a Tool in Evolutionary and Comparative Physiology: Insights into Complex Traits--an Introduction to the Symposium". Integrative and Comparative Biology. 45 (3): 387–390. doi:10.1093/icb/45.3.387. PMID 21676784. S2CID 2305227.
External links
[edit]- E. coli Long-term Experimental Evolution Project Site Archived 2017-07-27 at the Wayback Machine, Lenski lab, Michigan State University
- A movie illustrating the dramatic differences in wheel-running behavior.
- Experimental Evolution Publications by Ted Garland: Artificial Selection for High Voluntary Wheel-Running Behavior in House Mice — a detailed list of publications.
- Experimental Evolution — a list of laboratories that study experimental evolution.
- Network for Experimental Research on Evolution, University of California.
- Nicholls, Henry (30 September 2009). "My little zebra: The secrets of domestication". New Scientist.
- Inquiry-based middle school lesson plan: "Born to Run: Artificial Selection Lab"
- Digital Evolution for Education software


 French
French Deutsch
Deutsch