α-Aminoadipate pathway
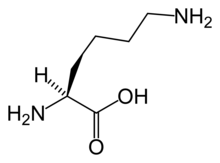
The α-aminoadipate pathway is a biochemical pathway for the synthesis of the amino acid L-lysine. In the eukaryotes, this pathway is unique to several species of yeast, higher fungi (containing chitin in their cell walls), and the euglenids.[1][2][3][4][5] It has also been reported from bacteria of the genus Thermus[6] and also in Pyrococcus horikoshii,[7] potentially suggesting a wider distribution than previously thought. This uniqueness of the pathway makes it a potentially interesting target for antimycotics.[3]
Pathway overview
[edit]
This pathway is a part of the glutamate family of amino acid biosynthetic pathways.[2] The reaction steps in the pathway are similar to the citric acid cycle.
The first step in the pathway is condensation of acetyl-CoA with α-ketoglutarate, which gives homocitrate. This reaction is catalyzed by homocitrate synthase. Homocitrate is then converted to homoaconitate by homoaconitase and then to homoisocitrate. This is then decarboxylated by homoisocitrate dehydrogenase, which results in α-ketoadipate. A nitrogen atom is added from glutamate by aminoadipate aminotransferase to form the α-aminoadipate, from which this pathway gets its name. This is then reduced by aminoadipate reductase via an acyl-enzyme intermediate to a semialdehyde. Reaction with glutamate by one class of saccharopine dehydrogenase yields saccharopine which is then cleaved by a second saccharopine dehydrogenase to yield lysine and oxoglutarate.[2]
Conversion of lysine to α-ketoadipate during degradation of lysine proceeds via the same steps, but in reverse.[8]
See also
[edit]References
[edit]- ^ Zabriskie TM, Jackson MD (2000). "Lysine biosynthesis and metabolism in fungi". Natural Product Reports. 17 (1): 85–97. doi:10.1039/a801345d. PMID 10714900.
- ^ a b c Xu H, Andi B, Qian J, West AH, Cook PF (2006). "The alpha-aminoadipate pathway for lysine biosynthesis in fungi". Cell Biochemistry and Biophysics. 46 (1): 43–64. doi:10.1385/CBB:46:1:43. PMID 16943623. S2CID 22370361.
- ^ a b Andi B, West AH, Cook PF (September 2004). "Kinetic mechanism of histidine-tagged homocitrate synthase from Saccharomyces cerevisiae". Biochemistry. 43 (37): 11790–11795. doi:10.1021/bi048766p. PMID 15362863.
- ^ Bhattacharjee JK (1985). "alpha-Aminoadipate pathway for the biosynthesis of lysine in lower eukaryotes". Critical Reviews in Microbiology. 12 (2): 131–151. doi:10.3109/10408418509104427. PMID 3928261.
- ^ Bhattacharjee JK, Strassman M (May 1967). "Accumulation of tricarboxylic acids related to lysine biosynthesis in a yeast mutant". The Journal of Biological Chemistry. 242 (10): 2542–2546. doi:10.1016/S0021-9258(18)95997-1. PMID 6026248.
- ^ Kosuge T, Hoshino T (1999). "The α-aminoadipate pathway for lysine biosynthesis is widely distributed among Thermus strains". Journal of Bioscience and Bioengineering. 88 (6): 672–5. doi:10.1016/S1389-1723(00)87099-1. PMID 16232683.
- ^ Nishida, Hiromi; Nishiyama, Makoto; Kobashi, Nobuyuki; Kosuge, Takehide; Hoshino, Takayuki; Yamane, Hisakazu (1999-12-01). "A Prokaryotic Gene Cluster Involved in Synthesis of Lysine through the Amino Adipate Pathway: A Key to the Evolution of Amino Acid Biosynthesis". Genome Research. 9 (12): 1175–1183. doi:10.1101/gr.9.12.1175. ISSN 1088-9051. PMID 10613839.
- ^ Voet, Donald; Voet, Judith G. (2011). Biochemistry (4. ed.). Hoboken, NJ: Wiley. ISBN 978-0-470-91745-9.


 French
French Deutsch
Deutsch