Cell cortex
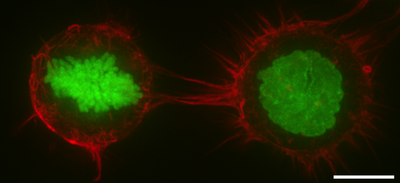
The cell cortex, also known as the actin cortex, cortical cytoskeleton or actomyosin cortex, is a specialized layer of cytoplasmic proteins on the inner face of the cell membrane. It functions as a modulator of membrane behavior and cell surface properties.[1][2][3] In most eukaryotic cells lacking a cell wall, the cortex is an actin-rich network consisting of F-actin filaments, myosin motors, and actin-binding proteins.[4][5] The actomyosin cortex is attached to the cell membrane via membrane-anchoring proteins called ERM proteins that plays a central role in cell shape control.[1][6] The protein constituents of the cortex undergo rapid turnover, making the cortex both mechanically rigid and highly plastic, two properties essential to its function. In most cases, the cortex is in the range of 100 to 1000 nanometers thick.
In some animal cells, the protein spectrin may be present in the cortex. Spectrin helps to create a network by cross-linked actin filaments.[3] The proportions of spectrin and actin vary with cell type.[7] Spectrin proteins and actin microfilaments are attached to transmembrane proteins by attachment proteins between them and the transmembrane proteins. The cell cortex is attached to the inner cytosolic face of the plasma membrane in cells where the spectrin proteins and actin microfilaments form a mesh-like structure that is continuously remodeled by polymerization, depolymerization and branching.
Many proteins are involved in the cortex regulation and dynamics, including formins, with roles in actin polymerization, Arp2/3 complexes that give rise to actin branching and capping proteins. Due to the branching process and the density of the actin cortex, the cortical cytoskeleton can comprise a highly complex meshwork such as a fractal structure.[8] Specialized cells are usually characterized by a very specific cortical actin cytoskeleton. For example, in red blood cells, the cell cortex consists of a two-dimensional cross-linked elastic network with pentagonal or hexagonal symmetry, tethered to the plasma membrane and formed primarily by spectrin, actin and ankyrin.[9] In neuronal axons, the actin or spectric cytoskeleton forms an array of periodic rings [10] and in the sperm flagellum it forms a helical structure.[11]
In plant cells, the cell cortex is reinforced by cortical microtubules underlying the plasma membrane. The direction of these cortical microtubules determines which way the cell elongates when it grows.
Functions
[edit]The cortex mainly functions to produce tension under the cell membrane, allowing the cell to change shape.[12] This is primarily accomplished through myosin II motors, which pull on the filaments to generate stress.[12] These changes in tension are required for the cell to change its shape as it undergoes cell migration and cell division.[12]
In mitosis, F-actin and myosin II form a highly contractile and uniform cortex to drive mitotic cell rounding. The surface tension produced by the actomyosin cortex activity generates intracellular hydrostatic pressure capable of displacing surrounding objects to facilitate rounding.[13][14] Thus, the cell cortex serves to protect the microtubule spindle from external mechanical disruption during mitosis.[15] When external forces are applied at sufficiently large rate and magnitude to a mitotic cell, loss of cortical F-actin homogeneity occurs leading to herniation of blebs and a temporary loss of the ability to protect the mitotic spindle.[16][17] Genetic studies have shown that the cell cortex in mitosis is regulated by diverse genes such as Rhoa,[18] WDR1,[19] ERM proteins,[20] Ect2,[21] Pbl, Cdc42, aPKC, Par6,[22] DJ-1 and FAM134A.[23]
In cytokinesis the cell cortex plays a central role by producing a myosin-rich contractile ring to constrict the dividing cell into two daughter cells.[24]
Cell cortex contractility is key for amoeboidal type cell migration characteristic of many cancer cell metastasis events.[1][25]
In addition to cell cortex also plays essential roles in the formation of tissues, organs and organisms. By pulling on adhesion complexes, the cortex promotes the expansion of contacts with other cells or with the extracellular matrix. Notably, during early mammalian development, the cortex pulls cells together to drive compaction and the formation of the morula.[26][27] Also, differences in cortical tension drives the sorting of the inner cell mass and trophectoderm progenitors during the formation of the morula,[28] the sorting of germ layer progenitors during zebrafish gastrulation,[29][30] the invagination of the mesoderm and the elongation of the germ band elongation during drosophila gastrulation.[31][32]
Research
[edit]Basic research into the cell cortex is done with immortalised cell lines, typically HeLa cells, S2 cells, Normal rat kidney cells, and M2 cells.[12] In M2 cells in particular, cellular blebs – which form without a cortex, then form one as they retract – are often used to model cortex formation and composition.[12]
References
[edit]- ^ a b c Salbreux G, Charras G, Paluch E (October 2012). "Actin cortex mechanics and cellular morphogenesis". Trends in Cell Biology. 22 (10): 536–45. doi:10.1016/j.tcb.2012.07.001. PMID 22871642.
- ^ Pesen D, Hoh JH (January 2005). "Micromechanical architecture of the endothelial cell cortex". Biophysical Journal. 88 (1): 670–9. Bibcode:2005BpJ....88..670P. doi:10.1529/biophysj.104.049965. PMC 1305044. PMID 15489304.
- ^ a b Alberts, Bruce; Johnson, Alexander; Lewis, Julian; Raff, Martin; Roberts, Keith; Walter, Peter (2002). "Cross-linking Proteins with Distinct Properties Organize Different Assemblies of Actin Filaments". Molecular Biology of the Cell (4th ed.). New York: Garland Science. ISBN 0-8153-3218-1.
- ^ Gunning PW, Ghoshdastider U, Whitaker S, Popp D, Robinson RC (June 2015). "The evolution of compositionally and functionally distinct actin filaments". Journal of Cell Science. 128 (11): 2009–19. doi:10.1242/jcs.165563. PMID 25788699.
- ^ Clark AG, Wartlick O, Salbreux G, Paluch EK (May 2014). "Stresses at the cell surface during animal cell morphogenesis". Current Biology. 24 (10): R484-94. Bibcode:2014CBio...24.R484C. doi:10.1016/j.cub.2014.03.059. PMID 24845681.
- ^ Fehon RG, McClatchey AI, Bretscher A (April 2010). "Organizing the cell cortex: the role of ERM proteins". Nature Reviews. Molecular Cell Biology. 11 (4): 276–87. doi:10.1038/nrm2866. PMC 2871950. PMID 20308985.
- ^ Machnicka B, Grochowalska R, Bogusławska DM, Sikorski AF, Lecomte MC (January 2012). "Spectrin-based skeleton as an actor in cell signaling". Cellular and Molecular Life Sciences. 69 (2): 191–201. doi:10.1007/s00018-011-0804-5. PMC 3249148. PMID 21877118.
- ^ Sadegh S, Higgins JL, Mannion PC, Tamkun MM, Krapf D (2017). "Plasma Membrane is Compartmentalized by a Self-Similar Cortical Actin Meshwork". Physical Review X. 7 (1): 011031. arXiv:1702.03997. Bibcode:2017PhRvX...7a1031S. doi:10.1103/PhysRevX.7.011031. PMC 5500227. PMID 28690919.
- ^ Gov NS (January 2007). "Active elastic network: cytoskeleton of the red blood cell". Physical Review E. 75 (1 Pt 1): 011921. Bibcode:2007PhRvE..75a1921G. doi:10.1103/PhysRevE.75.011921. PMID 17358198.
- ^ Xu K, Zhong G, Zhuang X (January 2013). "Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons". Science. 339 (6118): 452–6. Bibcode:2013Sci...339..452X. doi:10.1126/science.1232251. PMC 3815867. PMID 23239625.
- ^ Gervasi MG, Xu X, Carbajal-Gonzalez B, Buffone MG, Visconti PE, Krapf D (June 2018). "The actin cytoskeleton of the mouse sperm flagellum is organized in a helical structure". Journal of Cell Science. 131 (11): jcs215897. doi:10.1242/jcs.215897. PMC 6031324. PMID 29739876.
- ^ a b c d e Chugh P, Paluch EK (July 2018). "The actin cortex at a glance". J Cell Sci. 131 (14). doi:10.1242/jcs.186254. PMC 6080608. PMID 30026344.
- ^ Stewart MP, Helenius J, Toyoda Y, Ramanathan SP, Muller DJ, Hyman AA (January 2011). "Hydrostatic pressure and the actomyosin cortex drive mitotic cell rounding". Nature. 469 (7329): 226–30. Bibcode:2011Natur.469..226S. doi:10.1038/nature09642. PMID 21196934. S2CID 4425308.
- ^ Ramanathan SP, Helenius J, Stewart MP, Cattin CJ, Hyman AA, Muller DJ (February 2015). "Cdk1-dependent mitotic enrichment of cortical myosin II promotes cell rounding against confinement". Nature Cell Biology. 17 (2): 148–59. doi:10.1038/ncb3098. PMID 25621953. S2CID 5208968.
- ^ Lancaster, O (2013). "Mitotic Rounding Alters Cell Geometry to Ensure Efficient Bipolar Spindle Formation". Developmental Cell. 25 (3): 270–283. doi:10.1016/j.devcel.2013.03.014. PMID 23623611.
- ^ Charras, Guillaume; Paluch, Ewa (September 2008). "Blebs lead the way: how to migrate without lamellipodia". Nature Reviews Molecular Cell Biology. 9 (9): 730–736. doi:10.1038/nrm2453. PMID 18628785.
- ^ Cattin, Cedric (2015). "Mechanical control of mitotic progression in single animal cells". PNAS. 112 (36): 11258–11263. Bibcode:2015PNAS..11211258C. doi:10.1073/pnas.1502029112. PMC 4568679. PMID 26305930.
- ^ Maddox, A (2003). "RhoA is required for cortical retraction and rigidity during mitotic cell rounding". J. Cell Biol. 160 (2): 255–265. doi:10.1083/jcb.200207130. PMC 2172639. PMID 12538643. S2CID 1491406.
- ^ Fujibuchi, T (2005). "AIP1/WDR1 supports mitotic cell rounding". Biochem. Biophys. Res. Commun. 327 (1): 268–275. doi:10.1016/j.bbrc.2004.11.156. PMID 15629458.
- ^ Kunda, P (2008). "Moesin Controls Cortical Rigidity, Cell Rounding, and Spindle Morphogenesis during Mitosis". Current Biology. 18 (2): 91–101. Bibcode:2008CBio...18...91K. doi:10.1016/j.cub.2007.12.051. PMID 18207738. S2CID 831851.
- ^ Matthews, H (2013). "Changes in Ect2 Localization Couple Actomyosin-Dependent Cell Shape Changes to Mitotic Progression". Developmental Cell. 23 (2): 371–383. doi:10.1016/j.devcel.2012.06.003. PMC 3763371. PMID 22898780. S2CID 1295956.
- ^ Rosa, A (2015). "Ect2/Pbl acts via Rho and polarity proteins to direct the assembly of an isotropic actomyosin cortex upon mitotic entry". Developmental Cell. 32 (5): 604–616. doi:10.1016/j.devcel.2015.01.012. PMC 4359025. PMID 25703349. S2CID 17482918.
- ^ Toyoda, Y (2017). "Genome-scale single-cell mechanical phenotyping reveals disease-related genes involved in mitotic rounding". Nature Communications. 8 (1): 1266. Bibcode:2017NatCo...8.1266T. doi:10.1038/s41467-017-01147-6. PMC 5668354. PMID 29097687. S2CID 19567646.
- ^ Green RA, Paluch E, Oegema K (November 2012). "Cytokinesis in animal cells". Annual Review of Cell and Developmental Biology. 28: 29–58. doi:10.1146/annurev-cellbio-101011-155718. PMID 22804577.
- ^ Olson MF, Sahai E (April 2009). "The actin cytoskeleton in cancer cell motility". Clinical & Experimental Metastasis. 26 (4): 273–87. doi:10.1007/s10585-008-9174-2. PMID 18498004.
- ^ Maître, Jean-Léon; Niwayama, Ritsuya; Turlier, Hervé; Nédélec, François; Hiiragi, Takashi (July 2015). "Pulsatile cell-autonomous contractility drives compaction in the mouse embryo". Nature Cell Biology. 17 (7): 849–855. doi:10.1038/ncb3185. PMID 26075357.
- ^ Firmin, Julie; Ecker, Nicolas; Rivet Danon, Diane; Özgüç, Özge; Barraud Lange, Virginie; Turlier, Hervé; Patrat, Catherine; Maître, Jean-Léon (16 May 2024). "Mechanics of human embryo compaction". Nature. 629 (8012): 646–651. Bibcode:2024Natur.629..646F. doi:10.1038/s41586-024-07351-x. PMID 38693259.
- ^ Maître, Jean-Léon; Turlier, Hervé; Illukkumbura, Rukshala; Eismann, Björn; Niwayama, Ritsuya; Nédélec, François; Hiiragi, Takashi (August 2016). "Asymmetric division of contractile domains couples cell positioning and fate specification". Nature. 536 (7616): 344–348. Bibcode:2016Natur.536..344M. doi:10.1038/nature18958. PMC 4998956. PMID 27487217.
- ^ Krieg, M.; Arboleda-Estudillo, Y.; Puech, P.-H.; Käfer, J.; Graner, F.; Müller, D. J.; Heisenberg, C.-P. (April 2008). "Tensile forces govern germ-layer organization in zebrafish". Nature Cell Biology. 10 (4): 429–436. doi:10.1038/ncb1705. PMID 18364700.
- ^ Maître, Jean-Léon; Berthoumieux, Hélène; Krens, Simon Frederik Gabriel; Salbreux, Guillaume; Jülicher, Frank; Paluch, Ewa; Heisenberg, Carl-Philipp (12 October 2012). "Adhesion Functions in Cell Sorting by Mechanically Coupling the Cortices of Adhering Cells". Science. 338 (6104): 253–256. Bibcode:2012Sci...338..253M. doi:10.1126/science.1225399. PMID 22923438.
- ^ Bertet, Claire; Sulak, Lawrence; Lecuit, Thomas (June 2004). "Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation". Nature. 429 (6992): 667–671. Bibcode:2004Natur.429..667B. doi:10.1038/nature02590. PMID 15190355.
- ^ Martin, Adam C.; Kaschube, Matthias; Wieschaus, Eric F. (January 2009). "Pulsed contractions of an actin–myosin network drive apical constriction". Nature. 457 (7228): 495–499. Bibcode:2009Natur.457..495M. doi:10.1038/nature07522. PMC 2822715. PMID 19029882.
Further reading
[edit]- Bray, Dennis (2000). "Actin and Membranes". Cell Movements. pp. 81–101. doi:10.4324/9780203833582-7. ISBN 978-0-203-83358-2.


 French
French Deutsch
Deutsch