Phenylalanine—tRNA ligase
| Phenylalanine—tRNA ligase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 6.1.1.20 | ||||||||
| CAS no. | 9055-66-7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Ferredoxin-fold anticodon binding domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
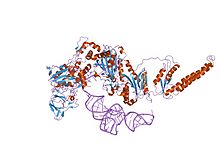 the crystal structure of phenylalanyl-tRNA synthetase from Thermus thermophilus complexed with cognate tRNAPhe | |||||||||
| Identifiers | |||||||||
| Symbol | FDX-ACB | ||||||||
| Pfam | PF03147 | ||||||||
| InterPro | IPR005121 | ||||||||
| SCOP2 | 1pys / SCOPe / SUPFAM | ||||||||
| |||||||||
In enzymology, a phenylalanine—tRNA ligase (EC 6.1.1.20) is an enzyme that catalyzes the chemical reaction
- ATP + L-phenylalanine + tRNAPhe AMP + diphosphate + L-phenylalanyl-tRNAPhe
The 3 substrates of this enzyme are ATP, L-phenylalanine, and tRNAPhe, whereas its 3 products are AMP, diphosphate, and L-phenylalanyl-tRNAPhe.
This enzyme belongs to the family of ligases, to be specific those forming carbon-oxygen bonds in aminoacyl-tRNA and related compounds. The systematic name of this enzyme class is L-phenylalanine:tRNAPhe ligase (AMP-forming). Other names in common use include phenylalanyl-tRNA synthetase, phenylalanyl-transfer ribonucleate synthetase, phenylalanine-tRNA synthetase, phenylalanyl-transfer RNA synthetase, phenylalanyl-tRNA ligase, phenylalanyl-transfer RNA ligase, L-phenylalanyl-tRNA synthetase, and phenylalanine translase. This enzyme participates in phenylalanine, tyrosine and tryptophan biosynthesis and aminoacyl-tRNA biosynthesis.
Phenylalanine-tRNA synthetase (PheRS) is known to be among the most complex enzymes of the aaRS (Aminoacyl-tRNA synthetase) family. Bacterial and mitochondrial PheRSs share a ferredoxin-fold anticodon binding (FDX-ACB) domain, which represents a canonical double split alpha+beta motif having no insertions. The FDX-ACB domain displays a typical RNA recognition fold (RRM) formed by the four-stranded antiparallel beta sheet, with two helices packed against it.[1][2][3][4][5]
Structural studies
[edit]As of late 2007, 10 structures have been solved for this class of enzymes, with PDB accession codes 1B70, 1B7Y, 1EIY, 1JJC, 1PYS, 2AKW, 2ALY, 2AMC, 2CXI, and 2IY5.
References
[edit]- ^ Mosyak L, Reshetnikova L, Goldgur Y, Delarue M, Safro MG (July 1995). "Structure of phenylalanyl-tRNA synthetase from Thermus thermophilus". Nat. Struct. Biol. 2 (7): 537–47. doi:10.1038/nsb0795-537. PMID 7664121. S2CID 13042127.
- ^ Goldgur Y, Mosyak L, Reshetnikova L, Ankilova V, Lavrik O, Khodyreva S, Safro M (January 1997). "The crystal structure of phenylalanyl-tRNA synthetase from thermus thermophilus complexed with cognate tRNAPhe". Structure. 5 (1): 59–68. doi:10.1016/s0969-2126(97)00166-4. PMID 9016717.
- ^ Rodova M, Ankilova V, Safro MG (February 1999). "Human phenylalanyl-tRNA synthetase: cloning, characterization of the deduced amino acid sequences in terms of the structural domains and coordinately regulated expression of the alpha and beta subunits in chronic myeloid leukemia cells". Biochem. Biophys. Res. Commun. 255 (3): 765–73. doi:10.1006/bbrc.1999.0141. PMID 10049785.
- ^ Moor N, Lavrik O, Favre A, Safro M (September 2003). "Prokaryotic and eukaryotic tetrameric phenylalanyl-tRNA synthetases display conservation of the binding mode of the tRNA(Phe) CCA end". Biochemistry. 42 (36): 10697–708. doi:10.1021/bi034732q. PMID 12962494.
- ^ Klipcan L, Levin I, Kessler N, Moor N, Finarov I, Safro M (July 2008). "The tRNA-induced conformational activation of human mitochondrial phenylalanyl-tRNA synthetase". Structure. 16 (7): 1095–104. doi:10.1016/j.str.2008.03.020. PMID 18611382.
Further reading
[edit]- Stulberg MP (1967). "The isolation and properties of phenylalanyl ribonucleic acid synthetase from Escherichia coli B". J. Biol. Chem. 242 (5): 1060–4. doi:10.1016/S0021-9258(18)96232-0. PMID 5335910.


 French
French Deutsch
Deutsch