Phylogenetic reconciliation

In phylogenetics, reconciliation is an approach to connect the history of two or more coevolving biological entities. The general idea of reconciliation is that a phylogenetic tree representing the evolution of an entity (e.g. homologous genes or symbionts) can be drawn within another phylogenetic tree representing an encompassing entity (respectively, species, hosts) to reveal their interdependence and the evolutionary events that have marked their shared history. The development of reconciliation approaches started in the 1980s, mainly to depict the coevolution of a gene and a genome, and of a host and a symbiont, which can be mutualist, commensalist or parasitic. It has also been used for example to detect horizontal gene transfer, or understand the dynamics of genome evolution.
Phylogenetic reconciliation can account for a diversity of evolutionary trajectories of what makes life's history, intertwined with each other at all scales that can be considered, from molecules to populations or cultures. A recent avatar of the importance of interactions between levels of organization is the holobiont concept, where a macro-organism is seen as a complex partnership of diverse species. Modeling the evolution of such complex entities is one of the challenging and exciting direction of current research on reconciliation.
Phylogenetic trees as nested structures
[edit]
Phylogenetic trees are intertwined at all levels of organization, integrating conflicts and dependencies within and between levels. Macro-organism populations migrate between continents, their microbe symbionts switch between populations, the genes of their symbionts transfer between microbe species, and domains are exchanged between genes. This list of organization levels is not representative or exhaustive, but gives a view of levels where reconciliation methods have been used. As a generic method, reconciliation could take into account numerous other levels. For instance, it could consider the syntenic organization of genes,[1] the interacting history of transposable elements and species,[2] the evolution of a protein complex across species.[3] The scale of evolutionary events considered can go from population events such as geographical diversification to nucleotids levels one inside genes,[4] including for instance chromosome levels events inside genomes such as whole genome duplication.[5]
Phylogenies have been used for representing the diversification of life at many levels of organization: macro-organisms,[6] their cells throughout development,[7] micro-organisms through marker genes,[8] chromosomes,[9] proteins,[10] protein domains,[6] and can also be helpful to understand the evolution of human culture elements such as languages[11] or fairy tales.[12] At each of these levels, phylogenetic trees describe different stories made of specific diversification events, which may or may not be shared among levels. Yet because they are structurally nested (similar to matryoshka dolls) or functionally dependent, the evolution at a particular level is bound to those at other levels.
Phylogenetic reconciliation is the identification of the links between levels through the comparison of at least two associated trees. Originally developed for two trees, reconciliations for more than two levels have been recently constructed (see section Explicit modeling of three or more levels). As such, reconciliation provides evolutionary scenarios that reveal conflict and cooperation among evolving entities. These links may be unintuitive, for instance, genes present in the same genome may show uncorrelated evolutionary histories while some genes present in the genome of a symbiont may show a strong coevolution signal with the host phylogeny. Hence, reconciliation can be a useful tool to understand the constraints and evolutionary strategies underlying the assemblage that forms a holobiont.
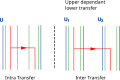
Because all levels essentially deal with the same object, a phylogenetic tree, the same models of reconciliation—in particular those based on duplication-transfer-loss events, which are central to this article—can be transposed, with slight modifications, to any pair of connected levels:[13] an "inner", "lower", or "associate" entity (e.g. gene, symbiont species, population) evolves inside an "upper", or "host" one (respectively species, host, or geographical area). The upper and lower entities are partially bound to the same history, leading to similarities in their phylogenetic trees, but the associations can change over time, become more or less strict or switch to other partners.
History
[edit]The principle of phylogenetic reconciliation was introduced in 1979[14] to account for differences between genes and species-level phylogenies. In a parsimonious setting, two evolutionary events, gene duplication and gene loss were invoked to explain the discrepancies between a gene tree and a species tree. It also described a score on gene trees knowing the species tree and an aligned sequence by using the number of gene duplication, loss, and nucleotide replacement for the evolution of the aligned sequence, an approach still central today with new models of reconciliation and phylogeny inference.[15]
The term reconciliation has been used by Wayne Maddison in 1997,[16] as a reverse concept of "phylogenetic discord" resulting from gene level evolutionary events.
Reconciliation was then developed jointly for the coevolution of host and symbiont and the geographic diversification of species. In both settings, it was important to model a horizontal event that implied parallel branches of the host tree: host switch for host and symbiont and species dispersion from one area to another in biogeography. Unlike for genes and genomes, the coevolution of host and symbiont and the explanation of species diversification by geography are not always the null hypothesis. A visual depiction of the two phylogenies in a tanglegram can help assess such coevolution, although it has no statistical obvious interpretation.[17]
Character methods, such as Brooks Parsimony Analysis,[18] were proposed to test coevolution and reconstruct scenarios of coevolution. In these methods, one of the trees is forgotten except for its leaves, which are then used as a character evolving on the second tree. First models for reconciliation, taking explicitly into account the two topologies and using a mechanistic event-based approach, were proposed for host and symbiont and biogeography.[19][20] Debates followed, as the methods were not yet completely sound but integrated useful information in a new framework.[21]
Costs for each event and a dynamic programming technique considering all pairs of host and symbiont nodes were then introduced into a host and symbiont approach, both of which still underlie most of the current reconciliation methods for host and symbiont as well as for species and genes.[22]
Reconciliation returned to the framework it was introduced in, gene and species. After character models were considered for horizontal gene transfer,[23] a new reconciliation model, following and improving the dynamic programming approach presented for host and symbiont, effectively introduced horizontal gene transfer to gene and species reconciliation on top of the duplication and loss model.[24]
The progressive development of phylogenetic reconciliation was thus possible through exchanges between multiple research communities studying phylogenies at the host and symbiont, gene and species, or biogeography levels. This story and its modern developments have been reviewed several times, generally focusing on specific pairs of levels, with a few exceptions.[25][13] New developments start to bring the different frameworks together with new integrative models.
Pocket Gophers and their chewing lices: a classical example
[edit]
Pocket gophers (Geomyidae) and their chewing lice (Trichodectidae) form a well studied system of host and symbiont coevolution.[26] The phylogeny of host and symbiont and the matching of the leaves of their trees are depicted on the left. For the host, O. stands for Orthogeomys, G. for Geomys and T. for Thomomys; for the symbiont, G. stands for Geomydoecus and T. for Thomoydoecus. Reconciling the two trees means giving a scenario with evolutionary events and matching on the ancestral nodes depicting the coevolution of the two trees. The events considered in this system are the events of the DTL model: duplication, transfer (or host switch), loss, and cospeciation, the null event of coevolution. Two scenarios were proposed in two studies,[27][28] using two different frameworks which could be deemed as pre-dynamic programming DTL[29] reconciliation. In modern DTL reconciliation frameworks, costs are assigned to events. The two scenarios were then shown to correspond to maximum parsimonious reconciliation with different cost assignments.[22] The scenario A uses 6 cospeciations, 2 duplications, 3 losses and 2 host switches to reconcile the two trees, while scenario B uses 5 cospeciations, 3 duplications, 3 losses and 2 host switches. The cost of a scenario is the sum of the cost of its events. For instance, with a cost of 0 for cospeciation, 2 for duplication, 1 for loss and 3 for host switch, scenario A has a cost of and scenario B of , and so according to a parsimonious principle, scenario A would be deemed more likely (scenario A stays more likely as long as the cost of cospeciation is less than the cost of duplication).
Development of Phylogenetic Reconciliation Models
[edit]
Models and methods used today in phylogeny are the result of several decades of research, made progressively complex, driven by the nature of the data and the quest for biological realism on one side, and the limits and progresses of mathematical and algorithmic methods on the other.
Pre-reconciliation models: characters on trees
[edit]Character methods can be used when there is no tree available for one of the levels, but only values for a character at the leaves of a phylogenetic tree for the other level. A model defines the events of character value change, their rate, probabilities or costs. For instance, the character can be the presence of a host on a symbiont tree,[18] the geographical region on a species tree,[31] the number of genes on a genome tree,[32] or nucleotides in a sequence.[4] Such methods thus aim at reconstructing ancestral characters at internal nodes of the tree.[33]
Although these methods have produced results on genome evolution, the utility of a second tree appears with very simple examples. If a symbiont has recently acquired the ability to spread in a group of species and thus it is present in most of them, character methods will wrongly indicate that the common ancestor of the hosts already had the symbiont. In contrast, a comparison of the symbiont and host trees would show discrepancies revealing horizontal transfers.
The origins of reconciliation: the Duplication Loss model and the Lowest Common Ancestor mapping
[edit]Duplication and loss were invoked first to explain the presence of multiple copies of a gene in a genome or its absence in certain species.[10] It is possible with those two events to reconcile any two trees,[14] i.e. to map the nodes and branches of the lower and upper trees, or equivalently to give a list of evolutionary events explaining the discrepancies between the upper tree and the lower tree. A most parsimonious Duplication and Loss (DL) reconciliation is computed through the Lowest Common Ancestor (LCA) mapping: proceeding from the leaves to the root, each internal node is mapped to the lowest common ancestor of the mapping of its two children.
A Markovian model for reconciliation
[edit]The LCA mapping in the DL model follows a parsimony principle: no event should be invoked if it is not necessary. However the use of this principle is debated,[4] and it is commonly admitted that it is more accurate in molecular evolution to fit a probabilistic model as a random walk, which does not necessarily produce parsimonious scenarios. A birth and death Markovian model is such a model that can generate a lower tree "inside" a fixed upper one from root to leaves.[34] Statistical inference provides a framework to find most likely scenarios, and in that case, a maximum likelihood reconciliation of two trees is also a parsimonious one. In addition, it is possible with such a framework to sample scenarios, or integrate over several possible scenarios in order to test different hypotheses, for example to explore the space of lower trees. Moreover, probabilistic models can be integrated into larger models, as probabilities simply multiply when assuming independence, for instance combining sequence evolution and DL reconciliation.[35]
Introducing horizontal transfer
[edit]
Host switch, i.e. inheritance of a symbiont from a kin lineage, is a crucial event in the evolution of parasitic or symbiotic relationships between species. This horizontal transfer also models migration events in biogeography and became of interest for the reconciliation of gene and species trees when it appeared that many discrepancies could not simply be explained by duplication and loss and that horizontal gene transfer (HGT) was a major evolutionary process in micro-organisms evolution. This switching, or horizontal transfer, pattern can also model admixture or introgression.[36] It is considered in character methods, without information from the symbiont phylogeny.[18][37] On top of the DL model, horizontal transfer enables new and very different reconciliation scenarios.
The simple yet powerful dynamic programming approach
[edit]The LCA reconciliation method yields a unique solution, which has been shown to be optimal for the problem of minimizing the weighted number of events, whatever the relative weights of duplication and loss.[38] In contrast, with Duplication, horizontal Transfer and Loss (DTL), there can be several equally parsimonious reconciliations. For instance, a succession of duplications and losses can be replaced by a single transfer. One of the first ideas to define a computational problem and approach a resolution was, in a host/symbiont framework, to maximize the number of co-speciations with a heuristic algorithm.[27] Another solution is to give relative costs to the events and find a scenario that minimizes the sum of the costs of its events.[22] In the probabilistic model frameworks, the equivalent task consists of assigning rates or probabilities to events and search for maximum likelihood scenarios, or sample scenarios according to their likelihood. All these problems are solved with a dynamic programming approach. This dynamic programming method involves traversing the two trees in a postorder. Proceeding from the leaves and then going up in the two trees, for each couple of internal nodes (one for each tree), the cost of a most parsimonious DTL reconciliation is computed.[22]
In a parsimony framework, costs of reconciling a lower subtree rooted at with an upper subtree rooted at is initialized for the leaves with their matching:
And then inductively, denoting the children of the children of the costs associated with speciation, duplication, horizontal transfer and loss, respectively (with often fixed to 0),
The costs and , because they do not depend on , can be computed once for all , hence achieving quadratic complexity to compute for all couples of and . The cost of losses only appears in association with other events because in parsimony, a loss can always be associated with the preceding event in the tree.
The induction behind the use of dynamic programming is based on always progressing in the trees toward the roots. However some combinations of events that can happen consecutively can make this induction ill-defined. One such combination consists of a transfer followed immediately by a loss in the donor lineage (TL). Restricting the use of this TL event repairs the induction.[39] With an unlimited use, it is necessary to use or add other known methods to solve systems of equations like fixed point methods,[40] or numerical solving of differential equations.[41] In 2016, only two out of seven of the most commonly used parsimony reconciliation programs did handle TL events,[42] although their consideration can drastically change the result of a reconciliation.[43]
Unlike LCA mapping, DTL reconciliation typically yields several scenarios of minimal cost, in some cases an exponential number. The strength of the dynamic programming approach is that it enables to compute a minimum cost of coevolution of the input upper and lower tree in quadratic time,[44] and to get a most parsimonious scenario through backtracking. It can also be transposed to a probabilistic framework to compute the likelihood of coevolution and get a most likely reconciliation, replacing costs with rates, minimums by sums and sums by products.[45] Moreover, through multiple backtracks, the approach is suitable for enumerating all parsimonious solutions or to sample scenarios, optimal and sub-optimal, according to their likelihood.
Estimation of event costs and rates
[edit]
Dynamic programming per se is only a partial solution and does not solve several problems raised by reconciliation. Defining a most parsimonious DTL reconciliation requires assigning costs to the different kinds of events (D, T and L). Different cost assignments can yield different reconciliation scenarios, so there is a need for a way to choose those costs. There is a diversity of approaches to do so. CoRe-PA[46] explores in a recursive manner the space of cost vectors, searching for a good matching with the event frequencies in reconciliations. ALE[45] uses the same idea in a probabilistic framework to estimate the event rates by maximum likelihood. Alternatively, COALA[47] is a preprocess using approximate Bayesian computation with sequential Monte Carlo: simulation and statistic rejection or acceptance of parameters with successive refinement.
In the parsimony framework, it is also possible to divide the space of possible event costs into areas of costs which lead to the same Pareto optimal solution.[48] Pareto optimal reconciliations are such that no other reconciliation has a strictly inferior cost for one type of event (duplication, transfer or loss), and less or equal for the others.
It is possible as well to rely on external considerations in order to choose the event costs. For example, the software Angst[49] chooses the costs that minimize the variation of genome size, in number of genes, between parent and children species.
The problem of temporal feasibility
[edit]
The dynamic programming method works for dated (internal nodes are totally ordered) or undated upper trees. However, with undated trees, there is a temporal feasibility issue. Indeed, a horizontal transfer implies that the donor and the receiver are contemporaneous, therefore implying a time constraint on the tree. In consequence, two horizontal transfers may be incompatible, because they imply contradicting time constraints. The dynamic programming approach can not easily check for such incompatibilities. If the upper tree is undated, finding a temporally feasible most parsimonious reconciliation is NP-hard.[50][51][52] It is fixed parameter tractable, which means that there are algorithms running in time bounded by an exponential of the number of transfers in the output scenarios.[51] Some solutions imply integer linear programming[53] or branch and bound exploration.[13] If the upper tree is dated, then there is no incompatibility issue because horizontal transfers can be constrained to never go backward in time. Finding a coherent optimal reconciliation is then solved in polynomial time[51] or with a speed-up in RASCAL,[54][55] by testing only a fraction of node mappings.
Most of the software taking undated trees does not look for temporal feasibility, except Jane,[56] which explores the space of total orders via a genetic algorithm, or, in a post process, Notung,[57] and Eucalypt,[58] which searches inside the set of optimal solutions for time consistent ones. Other methods work as supplementary layers to reconciliations, correcting reconciliations[59] or returning a subset of feasible transfers,[60] which can be used to date a species tree.[60][61]
Expanding phylogenies: Transfers from the dead
[edit]
In phylogenetics in general, it is important to keep in mind that the extant and ancestral species that are represented in any phylogeny are only a sparse sample of the species that currently exist or ever have existed. This is why one can safely assume that all transfers that can be detected using phylogenetic methods have originated in lineages that are, strictly speaking, absent from a studied phylogeny.[62] Accounting for extinct or unsampled biodiversity in phylogenetic studies can give a better understanding of these processes.[63] Originally, DTL reconciliation methods did not recognize this phenomenon and only allowed for transfer between contemporaneous branches of the tree, hence ignoring most plausible solutions. However, methods working on undated upper trees can be seen as implicitly handling the unknown diversity by allowing transfers "to the future" from the point of view of one phylogeny, that is, the donor is more ancient than the recipient. A transfer to the future can be translated into a speciation to unknown species, followed by a transfer from unknown species. ALE[62] in its dated version explicitly takes the unknown diversity into account by adding a Moran process of speciation/extinctions of species to the dated birth/death model of gene evolution. Transfers from the dead are also handled in a parsimonious setting by Tera and ecceTERA,[64][42] showing that considering these transfers improves the capacity to reconstruct gene trees using reconciliation, and with a more explicit model[65] and in a probabilistic setting, in ALE undated.[66]
The specificity of biogeography: a tree like structure for the "evolution" of areas
[edit]
In biogeography, some applications of reconciliation approaches consider as an upper tree an area cladogram with defined ancestral nodes. For instance, the root can be Pangaea and the nodes contemporary continents. Sometimes, internal nodes are not ancestral areas but the unions of the areas of their children, to account for the possibility of species evolving along the lower tree to inhabit one or several areas. In this case, the evolutionary events are migration, where one species colonizes a new area, allopatric speciation, or vicariance, equivalent to co-speciation in host/symbiont comparisons. Even though this approach does not always give a tree (if the unions AB and BC of leaves A, B, C exist, a child can have several parents), and this structure is not associated with time (it is possible for a species to go from A to AB by migration, as well as from AB to A by extinction), reconciliation methods—with events and dynamic programming—can infer evolutionary scenarios between the upper geographical structure and the lower species tree. Diva[67] and Lagrange[68][41] are two reconciliation models constructing such a tree-like structure and then applying reconciliation, the first with a parsimony principle, the second in a probabilistic framework. Additionally, BioGeoBEARS[69] is a biogeography inference package that reimplemented DIVA and Lagrange models and allows for new options, like distant dependent transfers[70] and discussion on statistical model selection.[69]
Graphical output
[edit]With two trees and multiple evolutionary events linking them to represent, viewing reconciled trees is a challenging but necessary question in order to make reconciliation studies more accessible. Some reconciliation software include annotation of the evolutionary events on the lower trees,[57] while others,[56][71][58][46] and specific packages, in DL[72] or DTL,[73] trace the lower tree embedded in the upper one. One difficulty in this regard is the variety of output formats for the different reconciliation software. A common standard, recphyloxml,[74] has been established and endorsed by part of the community, and a viewer is available, able to display reconciliation in multi level systems.[75]
Addressing Additional Practical Considerations
[edit]Applying DTL reconciliation to biological data raises several problems related to uncertainty and confidence levels of input and output. Concerning the output, the uncertainty of the answer calls for an exploration of the whole solution space. Concerning the input, phylogenetic reconciliation has to handle uncertainties in the resolution or rooting of the upper or lower trees, or even to propose roots or resolutions according to their confidence.
Exploring the space of reconciliations
[edit]
Dynamic programming makes it possible to sample reconciliations, uniformly among optimal ones[76] or according to their likelihood. It is also possible to enumerate them in time proportional to the number of solutions,[58] a number which can quickly become intractable (even only for optimal ones). Finding and presenting structure among the multitude of possible reconciliations has been at the center of recent methodological developments, especially for host and symbiont aimed methods. Several works have focused on representing a set of reconciliations in a compact way, from a uniform sample of optimal ones[76] or by constructing a graph summarizing the optimal solutions.[77] This can be achieved by giving support values to specific events based on all optimal (or suboptimal) reconciliations,[78] or with the use of a consensus reconciled tree.[79][59] In a DL model, it is possible to define a median reconciliation, based on shared events and to compute it in polynomial time.[80] EMPRess[71] can group similar reconciliations through clustering,[81] with all pairwise distance between reconciliations computable in polynomial time (independently of the number of most parsimonious reconciliations).[82] With the same aim, Capybara[83] defines equivalence classes among reconciliations, efficiently computing representatives for all classes, and outputs with linear delay a given number of reconciliations (first optimal ones, then sub optimal). The space of most parsimonious reconciliation can be expanded or reduced when increasing or decreasing horizontal transfer allowed distance,[58] which is easily done by dynamic programming.
Inferring phylogenetic trees with reconciliation
[edit]Reconciliation and input uncertainty
[edit]Reconciliation works with two fixed trees, a lower and an upper, both assumed correct and rooted. However, those trees are not first hand data. The most frequently used data for phylogenetics consists in aligned nucleotidic or proteic sequences. Extracting DNA, sequencing, assembling and annotating genomes, recognizing homology relationships among genes and producing multiple alignments for phylogenetic reconstruction are all complex processes where errors can ultimately affect the reconstructed tree.[84] Any topology or rooting error can be misinterpreted and cause systematic bias. For instance, in DL reconciliations, errors on the lower tree bias the reconciliation toward more duplication events closer to the root and more losses closer to the leaves.[85]
On the other hand, reconciliation, as a macro evolutionary model, can work as a supplementary layer to the micro evolutionary model of sequence evolution, resolving polytomies (nodes with more than two children) or rooting trees, or be intertwined with it through integrative models in order to get better phylogenies.
Most of the works in this direction focus on gene/species reconciliations, nevertheless some first steps have been made in host/symbiont, such as considering unrooted symbiont trees[86] or dealing with polytomies in Jane.[56]
Exploring the space of lower trees with reconciliation
[edit]Reconciliation can easily take unrooted lower trees as input, which is a frequently used feature because trees inferred from molecular data are typically unrooted. It is possible to test all possible roots, or a thoughtful triple traversal of the unrooted tree allows to do it without additional time complexity.[39] In a duplication-loss model, the set of roots minimizing the costs are found close to one another, forming a "plateau",[87] a property which does not generalize to DTL.[86][79]

Reconciliation can also take as input non binary trees, that is, with internal nodes with more than two children. Such trees can be obtained for example by contracting branches with low statistical support. Inferring a binary tree from a non binary tree according to reconciliation scores is solved in DL with efficient methods.[57][88][89][90][91] In DTL, the problem is NP hard.[92] Heuristics[93] and exact fixed parameter tractable algorithms[92][94][95] are possible solutions.
Another way to handle uncertainty in lower trees is to take as input a sample of alternative lower trees instead of a single one. For example, in the paper that gave reconciliation its name,[14] it was proposed to consider all most likely lower trees, and choose from these trees the best one according to their DL costs, a principle also used by TreeFix-DTL.[96] The sample of lower trees can similarly reflect their likelihood according to the aligned sequences, as obtained from Bayesian Markov chain Monte Carlo methods as implemented for example in Phylobayes.[97] AngST,[49] ALE[40] and ecceTERA[64] use "amalgamation", an extension of the DTL dynamic programming that is able to efficiently traverse a set of alternative lower trees instead of a single tree.
A local search in the space of lower trees guided by a joint likelihood, on the one hand from multiple sequence alignments and on the other hand from reconciliation with the upper tree, is achieved in Phyldog with a DL model[98] and in GeneRax with DTL.[15] In a DL model with sequence evolution and relaxed molecular clock, the lower tree space can be explored with an MCMC.[99] MowgliNNI[100] can modify the input gene tree at poorly supported nodes to increase DTL score, while TreeSolve resolves the multifurcations added by collapsing poorly supported nodes.[101]
Finally, integrative models—mixing sequence evolution and reconciliation—can compute a joint likelihood via dynamic programming (for both reconciliation and gene sequences evolution),[40] use Markov chain Monte Carlo to include molecular clock to estimate branch lengths, in a DL model[34] or with a relaxed molecular clock,[99] and in a DTL model.[102] These models have been applied in gene/species frameworks, not yet in host/symbiont or biogeography contexts.
Inferring upper trees using reconciliation
[edit]Inferring an upper tree from a set of lower trees is a long-standing question related to the supertree problem.[103] It is particularly interesting in the case of gene/species reconciliation where many (typically thousands of) gene trees are available from complete genome sequences. Supertree methods attempt to assemble a species tree based on sets of trees which may differ in terms of contemporary species sets and topology, but usually without consideration for the biological process explaining these differences. However, some supertree approaches are statistically consistent for the reconstruction of the species tree if the gene trees are simulated under a DL model. This means that if the number of input lower trees generated from the true upper tree via the DL model grows toward infinity, given that there are no additional errors, the output upper tree converges almost surely to the true one. This has been shown in the case of a quartet distance,[104] and with a generalised Robinson Foulds multicopy distance,[105][106] with better running time but assuming gene trees do not contain bipartitions contradicting the species tree, which seems rare under a DL model.

Reconciliation can also be used for the inference of upper trees. This is a computationally hard problem: already resolving polytomies in a non binary upper tree with a binary lower one—minimizing a DL reconciliation score—is NP-hard.[91] In particular, reconstructing the species tree giving the best DL cost for several gene trees is NP-hard and 2-approximable.[107] It is called the Gene Duplication problem or more generally Gene Tree parsimony. The problem was seen as a way to detect paralogy to get better species tree reconstruction.[108][109] It is NP-hard, with interesting results on the problem complexity[91][110] and the behaviour of the model with different input size, structure and ILS presence.[111] Multiple solutions exists, with ILP[112] or heuristics,[113][114] and with the possibility of a deep coalescence score.[115]
ODTL[45] takes as input gene trees and searches a maximum likelihood species tree according to a DTL model, with a hill-climbing search. The approach produces a species tree with internal nodes ordered in time, ensuring a time compatibility for the scenarios of transfer among lower trees {link section|The problem of temporal feasibility}.
Addressing a more general problem, Phyldog[98] searches for the maximum likelihood species tree, gene trees and DL parameters from multiple family alignments via multiple rounds of local search. It thus performs the exploration of both upper and lower trees at the same time. MixTreEM[116] presents a faster solution.
Limits of the two-level DTL model
[edit]A limit to dynamic programming: non independent evolution of children lineages
[edit]
The dynamic programming framework, like usual birth and death models, works under the hypothesis of independent evolution of children lineages in the lower tree. However, this hypothesis does not hold if the model is complemented with several other documented evolutionary events, such as horizontal transfer with replacement of a homologous gene in the recipient lineage, or gene conversion. Horizontal transfer with replacement is usually modeled by a rearrangement of the upper tree, called Subtree Prune and Regraft (SPR). Reconciling under SPR is NP-hard, even in dated trees, and fixed-parameter tractable regarding the output size.[117][118]
Another way to model and infer replacing horizontal transfers is through maximum agreement forest, where branches are cut in the lower and upper trees in order to get two identical (or statistically indistinguishable[119]) upper and lower forests. The problem is NP-hard,[120] but several approximations have been proposed.[121] Replacing transfers can be considered on top of the DL model.[122] In the same vein, gene conversion can be seen as a "replacing duplication". In this latter case, a polynomial algorithm which does not use dynamic programming and is an extension of the LCA method can find all optimal solutions, including gene conversions.[118]
Integrating population levels: failure to diverge and Incomplete Lineage Sorting
[edit]In host/symbiont frameworks, a single symbiont species is sometimes associated to several host species. This means that while a speciation or diversification has been observed in the host, the populations are indistinguishable in the symbiont. This is handled for example by additional polytomies in the symbiont tree, possibly leading to intractable inference problems, because polytomies need to be resolved. It is also modeled by an additional evolutionary event "failure to diverge" (Jane,[56] Amocoala[123]). Failure to diverge can be a way to allow "free" host switch in a population, a flow of symbionts between closely related hosts. Following that vision, host switch allowed only for close hosts is considered in Eucalypt.[58] This idea of horizontal flow between close populations can also be applied to gene/species frameworks, with a definition of species based on a gradient of gene flow between populations.[124]

Failure to diverge is one way of introducing population dynamics in reconciliation, a framework mainly adapted to the multi-species level, where populations are supposed to be well differentiated. There are other population phenomena that limit this framework, one of them being deep coalescence of lineages, leading to Incomplete Lineage Sorting (ILS), which is not handled by the DTL model.[88][125] The multi species coalescent is a classical model of allele evolution along a species tree, with birth of alleles and sorting of alleles at speciations, that takes into account population sizes and naturally encompasses ILS.[126][127][111][128][129] In a reconciliation context, several attempts have been made in order to account for ILS without the complex integration of a population model. For example, ILS can be seen as a possible evolutionary pattern for the gene tree. In that case, children lineages are not independent of one another, leading to intractability results. ILS alone can be handled with LCA, but ILS + DL reconciliation is NP hard, even without transfers.[130]
Notung[88] handles ILS by collapsing short branches of the species tree in polytomies and allowing ILS as a free diversification of gene trees on those polytomies. ecceTERA[131] binds the maximum size of connected parts of the species tree where ILS can happen, proposing a fixed parameter tractable algorithm in that parameter.
ILS and DL can be considered on an upper network instead of a tree. This models in particular introgression, with the possibility to estimate model parameters.[132]
More integrative reconciliation models accounting for ILS have been proposed, including both DL and multispecies coalescent,[133] with DLCoal. It is a probabilistic model with a parsimony translation,[134] proposing two sequential LCA-type heuristics handled via an intermediate locus tree between gene and species. However, outside of the gene/species reconciliation framework, ILS seems, for no particular reason, never considered in host/symbiont, nor in biogeography.
Cophylogeny with more than two levels
[edit]
A striking aspect of reconciliation is the common methodology handling different levels of organization: it is used for comparing domain and protein trees, gene and species trees, hosts and symbiont trees, population and geographic trees. However, now that scientists tend to consider that multi-level models of biological functioning bring a novel and game changing view of organisms and their environment,[136] the question is how to use reconciliation to bring phylogenetics to this holobiont era.
Coevolution of entities at different scales of evolution is at the basis of the holobiont idea: macro-organisms, micro-organisms and their genes all have a different history bound to a common functioning in a single ecosystem. Biological systems like the entanglement of host, symbionts and their genes imply functional and evolutionary dependencies between more than two levels.
Examples of multi level systems with complex evolutionary inter-dependencies
[edit]Genes coevolving beyond genome boundaries
[edit]The holobiont concept[137] stresses the possibility of genes from different genomes to cooperate and coevolve.[138][139][140] For instance, certain genes in a symbiont genome may provide a function to its host, like the production of a vital compound absent from available feeding sources. An iconic example is the case for blood-feeding or sap-feeding insects, which often depend on one or several bacterial symbionts to thrive on a resource that is abundant in sugar, but lacks essential amino-acids or vitamins.[141] Another example is the association of Fabaceae with nitrogen-fixing bacteria. The compound beneficiary to the host is typically produced by a set of genes encoded in the symbiont genome, which throughout evolution, may be transferred to other symbionts, and/or in and out of the host genome. Reconciliation methods have the potential to reveal evolutionary links between portions of genomes from different species. A search for coevolving genes beyond the boundaries of the genomes in which they are encoded would highlight the basis for the association of organisms in the holobiont.
Horizontal gene transfer routes depend on multiple levels
[edit]In intracellular mutualistic symbiont insect systems, multiple occurrences of horizontal gene transfers have been identified, whether from host to symbiont, symbiont to host or symbiont to symbiont.[142]
Transfers of endosymbiont genes involved in nutrition pathways beneficiary to the insect host have been shown to occur preferentially if the donor and recipient lineages share the same host.[143][144][145] This is also the case in insects with bacterial symbionts providing defensive protein[146] or in obligate leaf nodule bacterial symbionts associated with plants.[147] In the human host, gene transfer has been shown to occur preferentially among symbionts hosted in the same organs.[148]
A review of horizontal gene transfers in host/symbiont systems[149] stresses the importance of supporting HGTs with multiple evidence. Notably it is argued that transfers should be considered better supported when involving symbionts sharing a habitat, a geographical area, or the same host. One should, however, keep in mind that most of the diversity of hosts and symbionts is unknown and that transfers may have occurred in unsampled closely related species, hosts or symbionts.
The idea that gene transfer in symbionts is constrained by the host can also be used to investigate the host's phylogenetic history. For instance, based on phylogeographical studies, it is now accepted that the bacterium Helicobacter pylori has been associated with human populations since the origins of the human species.[150][151] An analysis of the genomes of Helicobacter pylori in Europe suggests that they are issued from a recombination between African and Asian Helicobacter pylori. This strongly implies early contacts between the corresponding human populations.
Similarly, an analysis of HGTs in coronaviruses from different mammalian species using reconciliation methods has revealed frequent contact between viral lineages, which can be interpreted as frequent host switches.[152]
Cultural evolution
[edit]The evolution of elements of human culture, for instance languages and folktales, in association with human population genetics, has been studied using concepts from phylogenetics. Although reconciliation has never been used in this framework, some of these studies encompass multiple levels of organization, each represented by a tree or the evolution of a character, with a focus on the coevolution of these levels.
Language trees can be compared with population trees in order to reveal vertically transmitted folktales, via a character model on this language tree.[153] Variants in each folktale's family, languages, genetic diversity, populations and geography can be compared two by two, to link folktale diversification with languages on one side and with geography on the other side.[154] As in genetics with symbionts sharing host promoting HGTs, linguistic barriers can foreclose the transmission of folktales or language elements.[155]
Investigating three-level systems using two-level reconciliation
[edit]Multi level reconciliation is not as developed as two-level reconciliation. One way to approach the evolutionary dependencies between more than two levels of organization is to try to use available standard two-level methods to give a first insight into a biological system's complexity.
- Multiple gene lineages can undergo joint events like segmental duplication, transfer or loss, or even whole genome duplication.
- Reconciliation can help identify highways of transfers and hybridizations.
- Three levels can be reconciled together, sequentially: the intermediate in the upper before adding the lower, or trying to find a joint most parsimonious scenario for the two reconciliations.
- With more than two levels, the reconciliation of the lower and intermediate levels can be compared to the reconciliation of the lower and upper.
Multi-gene events: implicit consideration of an intermediate level
[edit]At the gene/species tree level, one typically deals with many different gene trees. In this case, the hypothesis that different gene families evolve independently is made implicitly. However, this does not need to be the case. For instance, duplication, transfer and loss can occur for segments of a genome spanning an arbitrary number of contiguous genes. It is possible to consider such multi-gene events using an intermediate guide for lower trees inside the upper one. For instance, one can compute the joint likelihood of multiple gene tree reconciliations with a dated species tree with duplication, loss and whole genome duplication[5] or in a parsimonious setting,[156][157][158][159] and one definition of the problem is NP-hard.[160] Similarly, the DL framework can be enriched with duplication and loss of chromosome segments instead of a single gene. However, DL reconciliation becomes intractable with that new possibility.[161]
The link between two consecutive genes can also be modeled as an evolving character, subject to gain, loss, origination, breakage, duplication and transfer.[1] The evolution of this link appears as an additional level to species and gene trees, partly constrained by the gene/species tree reconciliation, partly evolving on its own, according to genome organization. It thus models the synteny, or proximity between genes. At another scale, it can as well model the evolution of two domains belonging to a protein.
The detection of "highways of transfers", the preferential acquisition of groups of genes from a specific donor, is another example of non-independence of gene histories.[162] Similarly, multi-gene transfers can be detected.[163] It has also led to methodological developments such as reconciliations using phylogenetic networks, seen as a tree augmented with transfer edges, which can be used to constrain transfers in a DTL model.[164] Networks can also be used to model introgression and incomplete lineage sorting.[165][166][36]
Detecting coevolution in multiple pairs of levels
[edit]It is a central question to understand the evolution of a holobiont to know what the levels are that coevolve with each other, for instance between host species, host genes, symbionts and symbiont genes. It is possible to approach the multiple inter-dependencies between all levels of evolution by multiple pairwise comparisons of two evolving entities.
Reconciliation of host and symbiont on one side and geography and symbiont on the other can also help to identify patterns of diversification of host and symbiont that reflect either coevolution or patterns that can be explained by a common geographical diversification.[167][168][169][170] Similarly, a study used reconciliation methods to differentiate the effect of diet evolution and phylogenetic inertia on the composition of mammalian gut microbiomes. By reconstructing ancestral diets and microbiome composition onto a mammalian phylogeny, the study revealed that both effects contribute but at different time scales.[171]
Explicit modeling of three or more levels
[edit]
In a model of a multi-level system as host/symbiont/genes, horizontal gene transfers should be more likely between two symbionts of a same host. This is invisible to a two-level gene tree/species tree or host/symbiont reconciliation: in some cases, looking at any combination of two levels can lead to missing an evolutionary scenario which can only be the most likely if the information from the three trees is considered together.
Trying to face the limitation of these uses of standard two-level reconciliations with systems involving inter-dependencies at multiple levels, a methodological effort has been undertaken in the last decade to construct and use multi-level models. This requires the identification of at least one "intermediate" level between the upper and the lower one.
- Two levels can be reconciled with the constraint of an upper one, for instance host and symbiont with geography.
- Characters can evolve on reconciled phylogenies, like gene synteny on a gene/species reconciliation.
- Transfers can be upper dependent, which is more likely between two intermediate entities that belong to a same upper one.
- With 3-levels reconciliation models, the intermediate tree can be inferred from the lower and upper trees.
Pre-reconciliation: characters onto reconciled trees
[edit]A first step towards integrated three-level models is to consider phylogenetic trees at two levels and another level represented only with characters at the leaves of one of the trees.
For instance, a reconciliation of host and symbiont phylogenies can be informed by geographic data.[172] Ancestral geographic locations of host and symbiont species obtained through a character inference method can then be used to constrain the host/symbiont reconciliation: ancestral hosts and symbionts can only be associated if they belong to the same geographical location.
At another scale, the evolution at the sub-gene level can be approached with a character method.[173] Here, parts of genes (e.g. the sequence coding for protein domains) is reconciled according to a DL model with a species tree, and the genes they belong to are mentioned as characters of these parts. Ancestral genes are then reconstructed a posteriori via merge and splits of gene parts.
Two-level reconciliations informed by a third level
[edit]As pointed out by several studies mentioned in § Horizontal transfer at multiple levels, an upper level can inform a reconciliation between an intermediate and lower one, notably for horizontal transfers.
Three-level models can take into account these assumptions to guide reconciliations between an intermediate tree and lower levels with the knowledge of an upper tree. The model can for example give higher likelihoods to reconciliation scenarios where horizontal gene transfers happen between entities sharing the same habitat. This has been achieved for the first time with DTL gene/species reconciliations nested with a DTL gene domain and gene reconciliation.[174] Different costs for inter and intra transfers depend on whether or not transfers happen between genes of the same genomes.
Note that this model explicitly considers three levels and three trees, but does not yet define a real three-level reconciliation, with a likelihood or score associated.[174] It relies on a sequential operation, where the second reconciliation is informed by the result of the first one.
The reconciliation problem in multi-level models
[edit]The next step is to define the score of a reconciliation consisting of three nested trees and to compute, given the three trees, three-level reconciliations according to their score. It has been achieved with a species/gene/domain system, where genes evolve within the species tree with a DL model and domains evolve within the gene/species system with a DTL model, forbidding domain transfers between genes of two different species.[175] Inference involves candidate scenarios with joint scores. Computing the minimum score scenario is NP-hard, but dynamic programming or integer linear programming can offer heuristics.[175][176] Variations of the problem considering multiple domains[177] are available, and so is a simulation framework.[178]
Inferring the intermediate tree using models of 3-level lower/intermediate/upper reconciliation
[edit]Just like two-level reconciliation can be used to improve lower or upper phylogenies, or to help constructing them from aligned sequences, joint reconciliation models can be used in the same manner.
In this vein, a coupled gene/species DL, domain gene DL and gene sequence evolution model in a Bayesian framework improves the reconstruction of gene trees.[179]
Software
[edit]Multiple pieces of software have been developed to implement the various models of reconciliation. The following table does not aim for exhaustiveness but presents a number of software tools aimed at reconciling trees to infer reconciliation scenarios or for related usage, such as correcting or inferring trees, or testing coevolution. The levels of interest section details the levels for which the software was implemented, even though it is entirely possible, for instance, to use a software made for species and gene reconciliation to reconcile host and symbionts.[180] Parsimony or probability is the underlying model that is used for the reconciliation.
| Name | Levels of interest | Platform | Command line or graphical user interface (GUI) | Usage | Probability or parsimony | Software availability and software license |
|---|---|---|---|---|---|---|
| Diva[181] | Geography and species | Unix, Mac, Win | Command line | Reconciliation inference | Parsimony | GNU GPLv2 |
| Lagrange[182] | Geography and species | Linux, Mac, Win | Python package | Reconciliation inference | Probability | GNU GPLv2 |
| BioGeoBEARS[183] | Geography and species | R package | R package | Reconciliation inference, statistical model test | Probability | GNU GPLv2, GNU GPLv3 |
| Jane[184] | Host and symbionts | Unix, Mac, Win | GUI or command line | Reconciliation inference, tree uncertainty | Parsimony | Proprietary, registration to download |
| eMPRess[185] | Host and symbionts | Unix, Mac, Win | GUI or command line | Reconciliation inference, cost estimation, solution space study | Parsimony | GNU GPLv3 |
| Eucalypt[186] | Host and symbionts | Java | Command line, graphical output with included viewer CophyTrees | Reconciliation inference, solution space study | Parsimony | CeCILL |
| Capybara[187] | Host and symbionts | Linux, Mac, Win, and Python package | GUI, Python package | Reconciliation inference, solution space study | Parsimony | code available on github |
| Coala[188] | Host and symbionts | Linux, Mac | Command line | cost estimation | Parsimony | CeCILL |
| RANGER-DTL[189] | Species and genes | Linux, Mac, Win | Command line | Reconciliation inference, tree uncertainty, solution sampling, replacing transfers | Parsimony | GNU GPLv3 |
| Notung[190] | Species and genes | Linux, Mac, Win < 7 | GUI | Reconciliation inference, tree uncertainty, gene, gene domain, species model | Parsimony | Proprietary |
| Mowgli[191] | Species and genes, Host and symbiont | Linux, Mac | Command line, graphical output with compatible viewer Sylvx | Reconciliation inference, tree uncertainty, ILS, geographical constraints, dated tree input | Parsimony | Proprietary |
| Sylvx[192] | Species and genes, Host and symbiont | Linux, Mac, Win | GUI | Viewer, compatible with Mowgli, ecceTERA | - | GNU GPLv2 or later |
| AnGST[193] | Species and genes | Python 2 | Command line | Reconciliation inference, cost estimation, dated tree input, tree uncertainty | Parsimony | source-available software[194] |
| ecceTERA[195] | Species and genes | Linux, Mac, built from code | Command line, compatible with Sylvx viewer, and recphyloxml output | Reconciliation inference, cost estimation, dated, partially dated or undated species tree input, tree uncertainty, reconciliation space study, species network | Parsimony | CeCILL |
| ALE[196] | Species and genes | Linux, Mac | Command line | Reconciliation inference, cost estimation, dated or undated species tree input, tree uncertainty | Probability | GNU GPLv3 |
| Treerecs[197] | Species and genes | Linux, Mac, Win | GUI, integrated to Seaview | DL tree correction | Parsimony, Probability | GNU Affero GPL Version 3.0-or-later |
| GeneRax[198] | Species and genes | Linux, Mac | Command line, graphical output with recphyloxml and thirdkind | Gene tree inference from reconciliation and aligned sequences, species tree inference | Probability | GNU Affero GPL v3.0 |
| Phyldog[199] | Species and genes | Linux, docker, vm | Command line | Gene and species tree inference from reconciliation and aligned sequences | Probability | CeCILL |
| CoRe-PA[200] | Host and symbiont | Linux, Mac, Win | GUI, Command line, graphical svg output | Reconciliation inference, cost estimation, dated tree, statistical test | Parsimony | - |
| CoRe-ILP[201] | Host and symbiont | Linux, Mac, Win | Command line | Reconciliation inference, temporal feasibility, dated tree | Parsimony | requires CPLEX (academic license can be obtained for free) |
| Rascal[55] | Host and symbiont | Java | Command line | Reconciliation inference, dated tree | Parsimony | - |
| MixTreEM[dead link] | Species and gene | Linux, Mac, Win (build from source) | Command line | Gene and species tree inference from reconciliation and aligned sequences | Probability | |
| PrIME[dead link] | Species and gene | Linux, Mac | Command line, graphical output (PrIMETV) | Reconciliation inference, gene and species tree inference from reconciliation and aligned sequences, orthology analysis | Probability | |
| Treemap[dead link] | Host and symbiont | Java | Command line | Reconciliation inference, statistical test | Parsimony | |
| JPrIME[202] | Gene and species | Java library | Command line | Reconciliation inference, gene and species tree inference from reconciliation and aligned sequences | Probability | New BSD |
| SEADOG[203] | Species and genes and domains | Linux, Mac | Command line | 3-level reconciliation inference | Parsimony | GNU GPLv3 |
| iGTP[204] | Species and genes | Linux, Mac, Win | GUI | Gene tree correction in DL or deep coalescence | Parsimony | Source code on request |
| TreeSolve[205] | Species and genes | Linux, Win | GUI | Gene tree correction in DTL | Parsimony | Source code on request |
| TreeFix,[206] TreeFix-DTL[207] | Species and genes | Linux | Command line | Gene tree correction in DL and DTL | Parsimony | GNU GPLv3 |
| ARTra[208] | Species and genes | Linux, Mac | Command line | Additive and replacing transfers inference | Parsimony | GNU GPL |
| DLCoal[209] | Species and genes | Linux, Mac, Win | Command line | Reconciliation inference with ILS | Parsimony | GNU GPL |
| Thirdkind[210] | Species and genes | Linux, Mac, Win | Command line | Viewer, compatible with recphyloxml | - | CeCILL |
References
[edit]![]() This article was adapted from the following source under a CC BY 4.0 license (2022) : Hugo Menet; Vincent Daubin; Eric Tannier (3 November 2022). "Phylogenetic reconciliation". PLOS Computational Biology. 18 (11): e1010621. doi:10.1371/JOURNAL.PCBI.1010621. ISSN 1553-734X. Wikidata Q115175616.
This article was adapted from the following source under a CC BY 4.0 license (2022) : Hugo Menet; Vincent Daubin; Eric Tannier (3 November 2022). "Phylogenetic reconciliation". PLOS Computational Biology. 18 (11): e1010621. doi:10.1371/JOURNAL.PCBI.1010621. ISSN 1553-734X. Wikidata Q115175616.
- ^ a b Duchemin, W.; Anselmetti, Y.; Patterson, M.; Ponty, Y.; Bérard, S.; Chauve, C.; Scornavacca, C.; Daubin, V.; Tannier, E. (2017). "DeCoSTAR: Reconstructing the Ancestral Organization of Genes or Genomes Using Reconciled Phylogenies". Genome Biology and Evolution. 9 (5): 1312–1319. doi:10.1093/gbe/evx069. PMC 5441342. PMID 28402423.
- ^ Loreto, E. L.; Carareto, C. M.; Capy, P. (2008). "Revisiting horizontal transfer of transposable elements in Drosophila". Heredity. 100 (6): 545–554. doi:10.1038/sj.hdy.6801094. PMID 18431403. S2CID 32424144.
- ^ Denise, R.; Abby, S. S.; Rocha EPC (2019). "Diversification of the type IV filament superfamily into machines for adhesion, protein secretion, DNA uptake, and motility". PLOS Biology. 17 (7): e3000390. doi:10.1371/journal.pbio.3000390. PMC 6668835. PMID 31323028.
- ^ a b c Felsenstein J (2004) Inferring Phylogenies. Oxford University Press
- ^ a b Zwaenepoel, A.; Van De Peer, Y. (2019). "Inference of Ancient Whole-Genome Duplications and the Evolution of Gene Duplication and Loss Rates". Molecular Biology and Evolution. 36 (7): 1384–1404. doi:10.1093/molbev/msz088. hdl:1854/LU-8612728. PMID 31004147.
- ^ a b Bagowski, C. P.; Bruins, W.; Te Velthuis, A. J. (2010). "The nature of protein domain evolution: Shaping the interaction network". Current Genomics. 11 (5): 368–376. doi:10.2174/138920210791616725. PMC 2945003. PMID 21286315.
- ^ Nair, N. U.; Lin, Y.; Manasovska, A.; Antic, J.; Grnarova, P.; Sahu, A. D.; Bucher, P.; Moret, B. M. (2014). "Study of cell differentiation by phylogenetic analysis using histone modification data". BMC Bioinformatics. 15 (1): 269. doi:10.1186/1471-2105-15-269. PMC 4138389. PMID 25104072.
- ^ Woese, C. R.; Kandler, O.; Wheelis, M. L. (1990). "Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya". Proceedings of the National Academy of Sciences of the United States of America. 87 (12): 4576–4579. Bibcode:1990PNAS...87.4576W. doi:10.1073/pnas.87.12.4576. PMC 54159. PMID 2112744.
- ^ Dobzhansky, T.; Sturtevant, A. H. (1938). "Inversions in the Chromosomes of Drosophila Pseudoobscura". Genetics. 23 (1): 28–64. doi:10.1093/genetics/23.1.28. PMC 1209001. PMID 17246876.
- ^ a b Zuckerkandl, E.; Pauling, L. (1965). "Molecules as documents of evolutionary history". Journal of Theoretical Biology. 8 (2): 357–366. Bibcode:1965JThBi...8..357Z. doi:10.1016/0022-5193(65)90083-4. PMID 5876245.
- ^ Gray, R. D.; Bryant, D.; Greenhill, S. J. (2010). "On the shape and fabric of human history". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 365 (1559): 3923–3933. doi:10.1098/rstb.2010.0162. PMC 2981918. PMID 21041216.
- ^ Tehrani, J. J. (2013). "The phylogeny of Little Red Riding Hood". PLOS ONE. 8 (11): e78871. Bibcode:2013PLoSO...878871T. doi:10.1371/journal.pone.0078871. PMC 3827309. PMID 24236061.
- ^ a b c Wieseke, Nicolas; Bernt, Matthias; Middendorf, Martin (30 July 2013). "Unifying Parsimonious Tree Reconciliation". arXiv:1307.7831 [q-bio.QM].
- ^ a b c Goodman, Morris; Czelusniak, John; Moore, G. William; Romero-Herrera, A. E.; Matsuda, Genji (1979). "Fitting the Gene Lineage into its Species Lineage, a Parsimony Strategy Illustrated by Cladograms Constructed from Globin Sequences". Systematic Zoology. 28 (2): 132–163. doi:10.2307/2412519. JSTOR 2412519.
- ^ a b Morel, B.; Kozlov, A. M.; Stamatakis, A.; Szöllősi, G. J. (2020). "GeneRax: A Tool for Species-Tree-Aware Maximum Likelihood-Based Gene Family Tree Inference under Gene Duplication, Transfer, and Loss". Molecular Biology and Evolution. 37 (9): 2763–2774. doi:10.1093/molbev/msaa141. PMC 8312565. PMID 32502238.
- ^ Wayne P. Maddison (1997) Gene Trees in Species Trees, Systematic Biology, 46(3) 523-536.
- ^ De Vienne, D. M. (2019). "Tanglegrams Are Misleading for Visual Evaluation of Tree Congruence". Molecular Biology and Evolution. 36 (1): 174–176. doi:10.1093/molbev/msy196. PMID 30351416.
- ^ a b c Brooks, Daniel R. (1981). "Hennig's Parasitological Method: A Proposed Solution". Systematic Zoology. 30 (3): 229–249. doi:10.2307/2413247. JSTOR 2413247.
- ^ Sneath, P. H. A. (1982). "Reviewed work: Systematics and Biogeography: Cladistics and Vicariance. II., Gareth Nelson, Norman Platnick". Systematic Zoology. 31 (2): 208–217. doi:10.2307/2413040. JSTOR 2413040.
- ^ Ronquist, Fredrik; Nylin, Soren (1990). "Process and Pattern in the Evolution of Species Associations". Systematic Zoology. 39 (4): 323–344. doi:10.2307/2992354. JSTOR 2992354.
- ^ Page RDM (1990). "Component Analysis: A Valiant Failure?". Cladistics. 6 (2): 119–136. doi:10.1111/j.1096-0031.1990.tb00532.x. PMID 34933509. S2CID 86307275.
- ^ a b c d Charleston, M. A. (1998). "Jungles: A new solution to the host/Parasite phylogeny reconciliation problem". Mathematical Biosciences. 149 (2): 191–223. doi:10.1016/s0025-5564(97)10012-8. PMID 9621683.
- ^ Hein, Jotun (1993). "A heuristic method to reconstruct the history of sequences subject to recombination". Journal of Molecular Evolution. 36 (4): 396. Bibcode:1993JMolE..36..396H. doi:10.1007/BF00182187. S2CID 16346458.
- ^ Hallett, M. T.; Lagergren, J. (2001). "Efficient algorithms for lateral gene transfer problems". Proceedings of the fifth annual international conference on Computational biology - RECOMB '01. pp. 149–156. doi:10.1145/369133.369188. ISBN 1581133537. S2CID 5804061.
- ^ Page, R. D. M. (1994). "Maps Between Trees and Cladistic Analysis of Historical Associations among Genes, Organisms, and Areas". Systematic Biology. 43: 58–77. doi:10.1093/sysbio/43.1.58.
- ^ Hafner, M. S.; Nadler, S. A. (1988). "Phylogenetic trees support the coevolution of parasites and their hosts". Nature. 332 (6161): 258–259. Bibcode:1988Natur.332..258H. doi:10.1038/332258a0. PMID 3347269. S2CID 4236760.
- ^ a b Page, Roderic D.M. (June 1994). "Parallel Phylogenies: Reconstructing the History of Host-Parasite Assemblages". Cladistics. 10 (2): 155–173. doi:10.1111/j.1096-0031.1994.tb00170.x.
- ^ Ronquist, F. (1995). "Reconstructing the History of Host-Parasite Associations Using Generalised Parsimony". Cladistics. 11 (1): 73–89. doi:10.1111/j.1096-0031.1995.tb00005.x. PMID 34920597. S2CID 86298930.
- ^ DTL stands for ""Duplication, horizontal Transfer and Loss
- ^ This table is intended to serve as illustration to the 2-Level reconciliation section of Menet et al. (2022) Phylogenetic reconciliation. PLoS Comput Biol 18(11): e1010621.doi:10.1371/journal.pcbi.1010621 and can be read along with it. Adding the horizontal Transfer event adds new, more parsimonious solutions compared to the previous DL model (A). With this new event, costs must be assigned to D, T and L events, and different costs give different solutions (B). Not all scenarios including transfers are time feasible. Some might include time constraints incompatible with the upper tree (C). Transfer can go from a species to one of its descendant via a sister lineages that went extinct (D). In biogeography, a tree like structure can be constructed to account for the possible migrations between different geographical areas (E). In some cases, an exponential number of scenarios might be most parsimonious, for example when two equivalent patterns have the same cost (F). The lower tree can be unrooted (G), multifurcating (H), or given as a sample of potential trees (I) and reconciliation can be used to resolve those uncertainties to get a binary rooted lower tree. Reconciliation score can also be used to help construct an upper tree (J). Dynamic programming is limited by the fact that it assumes independence between sister lineages, which makes it unable to consider replacing transfers or gene conversion (K), as well as Failure to diverge (L) and Incomplete Lineage Sorting (M), two population level events.
- ^ Wiley, E. O. (1988). "Parsimony Analysis and Vicariance Biogeography". Systematic Zoology. 37 (3): 271–290. doi:10.2307/2992373. JSTOR 2992373.
- ^ Csurös, M.; Miklós, I. (2009). "Streamlining and large ancestral genomes in Archaea inferred with a phylogenetic birth-and-death model". Molecular Biology and Evolution. 26 (9): 2087–2095. doi:10.1093/molbev/msp123. PMC 2726834. PMID 19570746.
- ^ Groussin, M.; Daubin, V.; Gouy, M.; Tannier, E. (2016). "Ancestral Reconstruction: Theory and Practice". Encyclopedia of Evolutionary Biology (PDF). pp. 70–77. doi:10.1016/B978-0-12-800049-6.00166-9. ISBN 9780128004265.
- ^ a b Arvestad, L.; Berglund, A. C.; Lagergren, J.; Sennblad, B. (2003). "Bayesian gene/Species tree reconciliation and orthology analysis using MCMC". Bioinformatics. 19 (Suppl 1): i7-15. doi:10.1093/bioinformatics/btg1000. PMID 12855432.
- ^ Arvestad, Lars; Berglund, Ann-Charlotte; Lagergren, Jens; Sennblad, Bengt (2004). "Gene tree reconstruction and orthology analysis based on an integrated model for duplications and sequence evolution". Proceedings of the eighth annual international conference on Computational molecular biology - RECOMB '04. pp. 326–335. doi:10.1145/974614.974657. ISBN 1581137559. S2CID 12364792.
- ^ a b Yu, Y.; Dong, J.; Liu, K. J.; Nakhleh, L. (2014). "Maximum likelihood inference of reticulate evolutionary histories". Proceedings of the National Academy of Sciences of the United States of America. 111 (46): 16448–16453. Bibcode:2014PNAS..11116448Y. doi:10.1073/pnas.1407950111. PMC 4246314. PMID 25368173.
- ^ Csurös, M. (2010). "Count: Evolutionary analysis of phylogenetic profiles with parsimony and likelihood". Bioinformatics. 26 (15): 1910–1912. doi:10.1093/bioinformatics/btq315. PMID 20551134.
- ^ Chauve C and El-Mabrouk N (2009) New Perspectives on Gene Family Evolution: Losses in Reconciliation and a Link with Supertrees. RECOMB 5541:46-58
- ^ a b Doyon J, Scornavacca C, Gorbunov K, Szöllősi G, Ranwez V et al. (2010) An Efficient Algorithm for Gene/Species Trees Parsimonious Reconciliation with Losses, Duplications and Transfers. RECOMB-CG
- ^ a b c Szöllõsi, G. J.; Rosikiewicz, W.; Boussau, B.; Tannier, E.; Daubin, V. (2013). "Efficient exploration of the space of reconciled gene trees". Systematic Biology. 62 (6): 901–912. doi:10.1093/sysbio/syt054. PMC 3797637. PMID 23925510.
- ^ a b Ree, R. H.; Smith, S. A. (2008). "Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis". Systematic Biology. 57 (1): 4–14. doi:10.1080/10635150701883881. PMID 18253896. S2CID 14205291.
- ^ a b Jacox, E.; Chauve, C.; Szöllősi, G. J.; Ponty, Y.; Scornavacca, C. (2016). "EcceTERA: Comprehensive gene tree-species tree reconciliation using parsimony". Bioinformatics. 32 (13): 2056–2058. doi:10.1093/bioinformatics/btw105. PMID 27153713.
- ^ Doyon, J. P.; Ranwez, V.; Daubin, V.; Berry, V. (2011). "Models, algorithms and programs for phylogeny reconciliation". Briefings in Bioinformatics. 12 (5): 392–400. doi:10.1093/bib/bbr045. PMID 21949266.
- ^ Bansal, M. S.; Alm, E. J.; Kellis, M. (2012). "Efficient algorithms for the reconciliation problem with gene duplication, horizontal transfer and loss". Bioinformatics. 28 (12): i283-91. doi:10.1093/bioinformatics/bts225. PMC 3371857. PMID 22689773.
- ^ a b c Szöllosi, G. J.; Boussau, B.; Abby, S. S.; Tannier, E.; Daubin, V. (2012). "Phylogenetic modeling of lateral gene transfer reconstructs the pattern and relative timing of speciations". Proceedings of the National Academy of Sciences of the United States of America. 109 (43): 17513–17518. Bibcode:2012PNAS..10917513S. doi:10.1073/pnas.1202997109. PMC 3491530. PMID 23043116.
- ^ a b Merkle, D.; Middendorf, M.; Wieseke, N. (2010). "A parameter-adaptive dynamic programming approach for inferring cophylogenies". BMC Bioinformatics. 11 (Suppl 1): S60. doi:10.1186/1471-2105-11-S1-S60. PMC 3009534. PMID 20122236.
- ^ Baudet, C.; Donati, B.; Sinaimeri, B.; Crescenzi, P.; Gautier, C.; Matias, C.; Sagot, M. F. (2015). "Cophylogeny reconstruction via an approximate Bayesian computation". Systematic Biology. 64 (3): 416–431. doi:10.1093/sysbio/syu129. PMC 4395844. PMID 25540454.
- ^ Libeskind-Hadas, R.; Wu, Y. C.; Bansal, M. S.; Kellis, M. (2014). "Pareto-optimal phylogenetic tree reconciliation". Bioinformatics. 30 (12): i87-95. doi:10.1093/bioinformatics/btu289. PMC 4058917. PMID 24932009.
- ^ a b David, L. A.; Alm, E. J. (2011). "Rapid evolutionary innovation during an Archaean genetic expansion". Nature. 469 (7328): 93–96. Bibcode:2011Natur.469...93D. doi:10.1038/nature09649. hdl:1721.1/61263. PMID 21170026. S2CID 4420725.
- ^ Hallett, M. T.; Lagergren, J. (2001). "Efficient algorithms for lateral gene transfer problems". Proceedings of the fifth annual international conference on Computational biology. pp. 149–156. doi:10.1145/369133.369188. ISBN 1581133537. S2CID 5804061.
- ^ a b c Tofigh, A.; Hallett, M.; Lagergren, J. (2011). "Simultaneous identification of duplications and lateral gene transfers". IEEE/ACM Transactions on Computational Biology and Bioinformatics. 8 (2): 517–535. doi:10.1109/TCBB.2010.14. PMID 21233529. S2CID 16462428.
- ^ Ovadia, Y.; Fielder, D.; Conow, C.; Libeskind-Hadas, R. (2011). "The co phylogeny reconstruction problem is NP-complete". Journal of Computational Biology. 18 (1): 59–65. doi:10.1089/cmb.2009.0240. PMID 20715926.
- ^ Wieseke, N.; Hartmann, T.; Bernt, M.; Middendorf, M. (2015). "Cophylogenetic Reconciliation with ILP". IEEE/ACM Transactions on Computational Biology and Bioinformatics. 12 (6): 1227–1235. doi:10.1109/TCBB.2015.2430336. PMID 26671795. S2CID 2291208.
- ^ Drinkwater, Benjamin; Charleston, Michael A. (2 January 2014). "An improved node mapping algorithm for the cophylogeny reconstruction problem". Coevolution. 2 (1): 1–17. doi:10.1080/23256214.2014.906070.
- ^ a b Drinkwater, B.; Charleston, M. A. (2016). "RASCAL: A Randomized Approach for Coevolutionary Analysis". Journal of Computational Biology. 23 (3): 218–227. doi:10.1089/cmb.2015.0111. PMID 26828619.
- ^ a b c d Conow, C.; Fielder, D.; Ovadia, Y.; Libeskind-Hadas, R. (2010). "Jane: A new tool for the cophylogeny reconstruction problem". Algorithms for Molecular Biology. 5: 16. doi:10.1186/1748-7188-5-16. PMC 2830923. PMID 20181081.
- ^ a b c Durand, D.; Halldórsson, B. V.; Vernot, B. (2006). "A hybrid micro-macroevolutionary approach to gene tree reconstruction". Journal of Computational Biology. 13 (2): 320–335. doi:10.1089/cmb.2006.13.320. PMID 16597243.
- ^ a b c d e Donati, B.; Baudet, C.; Sinaimeri, B.; Crescenzi, P.; Sagot, M. F. (2015). "EUCALYPT: Efficient tree reconciliation enumerator". Algorithms for Molecular Biology. 10 (1): 3. doi:10.1186/s13015-014-0031-3. PMC 4310143. PMID 25648467.
- ^ a b Ma, W.; Smirnov, D.; Forman, J.; Schweickart, A.; Slocum, C.; Srinivasan, S.; Libeskind-Hadas, R. (2018). "DTL-RNB: Algorithms and Tools for Summarizing the Space of DTL Reconciliations". IEEE/ACM Transactions on Computational Biology and Bioinformatics. 15 (2): 411–421. doi:10.1109/TCBB.2016.2537319. PMID 26955051. S2CID 4566350.
- ^ a b Chauve, Cédric; Rafiey, Akbar; Davín, Adrián A.; Scornavacca, Celine; Veber, Philippe; Boussau, Bastien; Szöllősi, Gergely J.; Daubin, Vincent; Tannier, Eric (14 April 2017). "MaxTiC: Fast ranking of a phylogenetic tree by Maximum Time Consistency with lateral gene transfers". doi:10.1101/127548. S2CID 21910535.
{{cite journal}}: Cite journal requires|journal=(help) - ^ Davín, A. A.; Tannier, E.; Williams, T. A.; Boussau, B.; Daubin, V.; Szöllősi, G. J. (2018). "Gene transfers can date the tree of life". Nature Ecology & Evolution. 2 (5): 904–909. Bibcode:2018NatEE...2..904D. doi:10.1038/s41559-018-0525-3. PMC 5912509. PMID 29610471.
- ^ a b Szöllosi, G. J.; Tannier, E.; Lartillot, N.; Daubin, V. (2013). "Lateral gene transfer from the dead". Systematic Biology. 62 (3): 386–397. doi:10.1093/sysbio/syt003. PMC 3622898. PMID 23355531.
- ^ Davín, A. A.; Tricou, T.; Tannier, E.; De Vienne, D. M.; Szöllősi, G. J. (2020). "Zombi: A phylogenetic simulator of trees, genomes and sequences that accounts for dead linages". Bioinformatics. 36 (4): 1286–1288. doi:10.1093/bioinformatics/btz710. PMC 7031779. PMID 31566657.
- ^ Weiner, Samson; Bansal, Mukul S. (5 August 2021). "Improved Duplication-Transfer-Loss Reconciliation with Extinct and Unsampled Lineages". Algorithms. 14 (8): 231. doi:10.3390/a14080231.
- ^ Szöllősi, G. J.; Davín, A. A.; Tannier, E.; Daubin, V.; Boussau, B. (2015). "Genome-scale phylogenetic analysis finds extensive gene transfer among fungi". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 370 (1678). doi:10.1098/rstb.2014.0335. PMC 4571573. PMID 26323765.
- ^ Ronquist F (1997) Phylogenetic approaches in coevolution and biogeography. Zoologica Scripta 26:313--322
- ^ Ree, R. H.; Moore, B. R.; Webb, C. O.; Donoghue, M. J. (2005). "A likelihood framework for inferring the evolution of geographic range on phylogenetic trees". Evolution; International Journal of Organic Evolution. 59 (11): 2299–2511. doi:10.1111/j.0014-3820.2005.tb00940.x. PMID 16396171. S2CID 23245573.
- ^ a b Matzke, N. J. (2014). "Model selection in historical biogeography reveals that founder-event speciation is a crucial process in Island Clades". Systematic Biology. 63 (6): 951–970. doi:10.1093/sysbio/syu056. PMID 25123369.
- ^ Van Dam, Matthew H.; Matzke, Nicholas J. (2016). "Evaluating the influence of connectivity and distance on biogeographical patterns in the south-western deserts of North America". Journal of Biogeography. 43 (8): 1514–1532. Bibcode:2016JBiog..43.1514V. doi:10.1111/jbi.12727. S2CID 87276317.
- ^ a b Santichaivekin, S.; Yang, Q.; Liu, J.; Mawhorter, R.; Jiang, J.; Wesley, T.; Wu, Y. C.; Libeskind-Hadas, R. (2021). "EMPRess: A systematic cophylogeny reconciliation tool". Bioinformatics. 37 (16): 2481–2482. doi:10.1093/bioinformatics/btaa978. PMID 33216126.
- ^ Sennblad, B.; Schreil, E.; Berglund Sonnhammer, A. C.; Lagergren, J.; Arvestad, L. (2007). "Primetv: A viewer for reconciled trees". BMC Bioinformatics. 8: 148. doi:10.1186/1471-2105-8-148. PMC 1891116. PMID 17484781.
- ^ Chevenet, F.; Doyon, J. P.; Scornavacca, C.; Jacox, E.; Jousselin, E.; Berry, V. (2016). "SylvX: A viewer for phylogenetic tree reconciliations". Bioinformatics. 32 (4): 608–610. doi:10.1093/bioinformatics/btv625. PMID 26515823.
- ^ Duchemin, W.; Gence, G.; Arigon Chifolleau, A. M.; Arvestad, L.; Bansal, M. S.; Berry, V.; Boussau, B.; Chevenet, F.; Comte, N.; Davín, A. A.; Dessimoz, C.; Dylus, D.; Hasic, D.; Mallo, D.; Planel, R.; Posada, D.; Scornavacca, C.; Szöllosi, G.; Zhang, L.; Daubin, V. (2018). "RecPhyloXML: A format for reconciled gene trees". Bioinformatics. 34 (21): 3646–3652. doi:10.1093/bioinformatics/bty389. PMC 6198865. PMID 29762653.
- ^ Penel, S.; Menet, H.; Tricou, T.; Daubin, V.; Tannier, E. (2022). "Thirdkind: Displaying phylogenetic encounters beyond 2-level reconciliation". Bioinformatics. 38 (8): 2350–2352. doi:10.1093/bioinformatics/btac062. PMID 35139153.
- ^ a b Bansal, M. S.; Alm, E. J.; Kellis, M. (2013). "Reconciliation revisited: Handling multiple optima when reconciling with duplication, transfer, and loss". Journal of Computational Biology. 20 (10): 738–754. doi:10.1089/cmb.2013.0073. PMC 3791060. PMID 24033262.
- ^ Nguyen, T. H.; Ranwez, V.; Berry, V.; Scornavacca, C. (2013). "Support measures to estimate the reliability of evolutionary events predicted by reconciliation methods". PLOS ONE. 8 (10): e73667. Bibcode:2013PLoSO...873667N. doi:10.1371/journal.pone.0073667. PMC 3790797. PMID 24124449.
- ^ a b Kundu, S.; Bansal, M. S. (2018). "On the impact of uncertain gene tree rooting on duplication-transfer-loss reconciliation". BMC Bioinformatics. 19 (Suppl 9): 290. doi:10.1186/s12859-018-2269-0. PMC 6101088. PMID 30367593.
- ^ Huber K, Moulton V, Sagot M-F, Sinaimeri B (2018) Geometric medians in reconciliation spaces of phylogenetic trees. Information Processing Letter. 136: 96–101
- ^ Mawhorter, R.; Libeskind-Hadas, R. (2019). "Hierarchical clustering of maximum parsimony reconciliations". BMC Bioinformatics. 20 (1): 612. doi:10.1186/s12859-019-3223-5. PMC 6882150. PMID 31775628.
- ^ Santichaivekin, S.; Mawhorter, R.; Libeskind-Hadas, R. (2019). "An efficient exact algorithm for computing all pairwise distances between reconciliations in the duplication-transfer-loss model". BMC Bioinformatics. 20 (Suppl 20): 636. doi:10.1186/s12859-019-3203-9. PMC 6915856. PMID 31842734.
- ^ Wang, Y.; Mary, A.; Sagot, M. F.; Sinaimeri, B. (2020). "Capybara: Equivalence ClAss enumeration of coPhylogenY event-BAsed ReconciliAtions". Bioinformatics. 36 (14): 4197–4199. doi:10.1093/bioinformatics/btaa498. PMID 32556075.
- ^ Boussau, B.; Daubin, V. (2010). "Genomes as documents of evolutionary history". Trends in Ecology & Evolution. 25 (4): 224–232. Bibcode:2010TEcoE..25..224B. doi:10.1016/j.tree.2009.09.007. PMID 19880211.
- ^ Hahn, M. W. (2007). "Bias in phylogenetic tree reconciliation methods: Implications for vertebrate genome evolution". Genome Biology. 8 (7): R141. doi:10.1186/gb-2007-8-7-r141. PMC 2323230. PMID 17634151.
- ^ a b Urbini, L.; Sinaimeri, B.; Matias, C.; Sagot, M. F. (2018). "Exploring the Robustness of the Parsimonious Reconciliation Method in Host-Symbiont Cophylogeny" (PDF). IEEE/ACM Transactions on Computational Biology and Bioinformatics. 16 (3): 738–748. doi:10.1109/TCBB.2018.2838667. PMID 29993554. S2CID 51613681.
- ^ Górecki, P.; Eulenstein, O.; Tiuryn, J. (2013). "Unrooted tree reconciliation: A unified approach". IEEE/ACM Transactions on Computational Biology and Bioinformatics. 10 (2): 522–536. doi:10.1109/TCBB.2013.22. PMID 23929875. S2CID 13416908.
- ^ a b c Stolzer, M.; Lai, H.; Xu, M.; Sathaye, D.; Vernot, B.; Durand, D. (2012). "Inferring duplications, losses, transfers and incomplete lineage sorting with nonbinary species trees". Bioinformatics. 28 (18): i409 – i415. doi:10.1093/bioinformatics/bts386. PMC 3436813. PMID 22962460.
- ^ Lafond M and Noutahi E and El-Mabrouk N (2016) Efficient Non-Binary Gene Tree Resolution with Weighted Reconciliation Cost. 27th Annual Symposium on Combinatorial Pattern Matching (CPM 2016) 14:1--14:12
- ^ Comte, N.; Morel, B.; Hasić, D.; Guéguen, L.; Boussau, B.; Daubin, V.; Penel, S.; Scornavacca, C.; Gouy, M.; Stamatakis, A.; Tannier, E.; Parsons, D. P. (2020). "Treerecs: An integrated phylogenetic tool, from sequences to reconciliations". Bioinformatics. 36 (18): 4822–4824. doi:10.1093/bioinformatics/btaa615. PMID 33085745.
- ^ a b c Zheng, Yu; Wu, Taoyang; Zhang, Louxin (2012). "Reconciliation of Gene and Species Trees with Polytomies". arXiv:1201.3995 [q-bio.PE].
- ^ a b Kordi, M.; Bansal, M. S. (2017). "On the Complexity of Duplication-Transfer-Loss Reconciliation with Non-Binary Gene Trees". IEEE/ACM Transactions on Computational Biology and Bioinformatics. 14 (3): 587–599. doi:10.1109/TCBB.2015.2511761. PMID 28055898. S2CID 4458502.
- ^ Lai, Han; Stolzer, Maureen; Durand, Dannie (2017). "Fast Heuristics for Resolving Weakly Supported Branches Using Duplication, Transfers, and Losses". Comparative Genomics. Lecture Notes in Computer Science. Vol. 10562. pp. 298–320. doi:10.1007/978-3-319-67979-2_16. ISBN 978-3-319-67978-5.
- ^ Jacox, E.; Weller, M.; Tannier, E.; Scornavacca, C. (2017). "Resolution and reconciliation of non-binary gene trees with transfers, duplications and losses". Bioinformatics. 33 (7): 980–987. doi:10.1093/bioinformatics/btw778. PMID 28073758.
- ^ Kordi, M.; Bansal, M. S. (2019). "Exact Algorithms for Duplication-Transfer-Loss Reconciliation with Non-Binary Gene Trees". IEEE/ACM Transactions on Computational Biology and Bioinformatics. 16 (4): 1077–1090. doi:10.1109/TCBB.2017.2710342. PMID 28622673. S2CID 54606569.
- ^ Bansal, M. S.; Wu, Y. C.; Alm, E. J.; Kellis, M. (2015). "Improved gene tree error correction in the presence of horizontal gene transfer". Bioinformatics. 31 (8): 1211–1218. doi:10.1093/bioinformatics/btu806. PMC 4393519. PMID 25481006.
- ^ Lartillot, N.; Philippe, H. (2004). "A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process". Molecular Biology and Evolution. 21 (6): 1095–1109. doi:10.1093/molbev/msh112. PMID 15014145.
- ^ a b Boussau, B.; Szöllosi, G. J.; Duret, L.; Gouy, M.; Tannier, E.; Daubin, V. (2013). "Genome-scale coestimation of species and gene trees". Genome Research. 23 (2): 323–330. doi:10.1101/gr.141978.112. PMC 3561873. PMID 23132911.
- ^ a b Akerborg, O.; Sennblad, B.; Arvestad, L.; Lagergren, J. (2009). "Simultaneous Bayesian gene tree reconstruction and reconciliation analysis". Proceedings of the National Academy of Sciences of the United States of America. 106 (14): 5714–5719. Bibcode:2009PNAS..106.5714A. doi:10.1073/pnas.0806251106. PMC 2667006. PMID 19299507.
- ^ Nguyen, T. H.; Ranwez, V.; Pointet, S.; Chifolleau, A. M.; Doyon, J. P.; Berry, V. (2013). "Reconciliation and local gene tree rearrangement can be of mutual profit". Algorithms for Molecular Biology. 8 (1): 12. doi:10.1186/1748-7188-8-12. PMC 3871789. PMID 23566548.
- ^ Kordi, Misagh; Bansal, Mukul S. (2020). "Tree Solve: Rapid Error-Correction of Microbial Gene Trees". Algorithms for Computational Biology. Lecture Notes in Computer Science. Vol. 12099. pp. 125–139. doi:10.1007/978-3-030-42266-0_10. ISBN 978-3-030-42265-3. S2CID 210171366.
- ^ Sjöstrand, J.; Tofigh, A.; Daubin, V.; Arvestad, L.; Sennblad, B.; Lagergren, J. (2014). "A Bayesian method for analyzing lateral gene transfer". Systematic Biology. 63 (3): 409–420. doi:10.1093/sysbio/syu007. PMID 24562812.
- ^ Warnow, Tandy (2018). "Supertree Construction: Opportunities and Challenges". arXiv:1805.03530 [q-bio.PE].
- ^ Legried, B.; Molloy, E. K.; Warnow, T.; Roch, S. (2021). "Polynomial-Time Statistical Estimation of Species Trees Under Gene Duplication and Loss". Journal of Computational Biology. 28 (5): 452–468. doi:10.1089/cmb.2020.0424. PMID 33325781.
- ^ Molloy, E. K.; Warnow, T. (2020). "FastMulRFS: Fast and accurate species tree estimation under generic gene duplication and loss models". Bioinformatics. 36 (Suppl_1): i57 – i65. doi:10.1093/bioinformatics/btaa444. PMC 7355287. PMID 32657396.
- ^ Chaudhary, R.; Burleigh, J. G.; Fernández-Baca, D. (2013). "Inferring species trees from incongruent multi-copy gene trees using the Robinson-Foulds distance". Algorithms for Molecular Biology. 8 (1): 28. doi:10.1186/1748-7188-8-28. PMC 3874668. PMID 24180377.
- ^ Ma B and Li M and Zhang L (2000) From Gene Trees to Species Trees. SIAM J. Comput., may, 729–752 24
- ^ Guigo, Roderic; Muchnik, Ilya; Smith, Temple F. (1 October 1996). "Reconstruction of Ancient Molecular Phylogeny". Molecular Phylogenetics and Evolution. 6 (2): 189–213. Bibcode:1996MolPE...6..189G. doi:10.1006/mpev.1996.0071. ISSN 1055-7903. PMID 8899723.
- ^ Page, Roderic D. M. (1 January 2000). "Extracting Species Trees From Complex Gene Trees: Reconciled Trees And Vertebrate Phylogeny". Molecular Phylogenetics and Evolution. 14 (1): 89–106. Bibcode:2000MolPE..14...89P. doi:10.1006/mpev.1999.0676. ISSN 1055-7903. PMID 10631044. Retrieved 17 December 2022.
- ^ Bansal, M. S.; Shamir, R. (2011). "A Note on the Fixed Parameter Tractability of the Gene-Duplication Problem". IEEE/ACM Transactions on Computational Biology and Bioinformatics. 8 (3): 848–850. doi:10.1109/TCBB.2010.74. PMID 20733245. S2CID 7086924.
- ^ a b Maddison, W. P.; Knowles, L. L. (2006). "Inferring phylogeny despite incomplete lineage sorting". Systematic Biology. 55 (1): 21–30. doi:10.1080/10635150500354928. PMID 16507521. S2CID 13453831.
- ^ Chang, W. C.; Burleigh, G. J.; Fernández-Baca, D. F.; Eulenstein, O. (2011). "An ILP solution for the gene duplication problem". BMC Bioinformatics. 12 (Suppl 1): S14. doi:10.1186/1471-2105-12-S1-S14. PMC 3044268. PMID 21342543.
- ^ Page, R. D. (1998). "GeneTree: Comparing gene and species phylogenies using reconciled trees". Bioinformatics. 14 (9): 819–820. doi:10.1093/bioinformatics/14.9.819. PMID 9918954.
- ^ Wehe, A.; Bansal, M. S.; Burleigh, J. G.; Eulenstein, O. (2008). "DupTree: A program for large-scale phylogenetic analyses using gene tree parsimony". Bioinformatics. 24 (13): 1540–1541. doi:10.1093/bioinformatics/btn230. PMID 18474508.
- ^ Chaudhary, R.; Bansal, M. S.; Wehe, A.; Fernández-Baca, D.; Eulenstein, O. (2010). "IGTP: A software package for large-scale gene tree parsimony analysis". BMC Bioinformatics. 11: 574. doi:10.1186/1471-2105-11-574. PMC 3002902. PMID 21092314.
- ^ Ullah, I.; Parviainen, P.; Lagergren, J. (2015). "Species Tree Inference Using a Mixture Model". Molecular Biology and Evolution. 32 (9): 2469–2482. doi:10.1093/molbev/msv115. PMID 25963975.
- ^ Bordewich W and Semple C (2005) On the Computational Complexity of the Rooted Subtree Prune and Regraft Distance. Annals of Combinatoris 8: 409-423
- ^ a b Hasić, D.; Tannier, E. (2019). "Gene tree species tree reconciliation with gene conversion". Journal of Mathematical Biology. 78 (6): 1981–2014. arXiv:1703.08950. doi:10.1007/s00285-019-01331-w. PMID 30767052. S2CID 8476555.
- ^ Abby, S. S.; Tannier, E.; Gouy, M.; Daubin, V. (2010). "Detecting lateral gene transfers by statistical reconciliation of phylogenetic forests". BMC Bioinformatics. 11: 324. doi:10.1186/1471-2105-11-324. PMC 2905365. PMID 20550700.
- ^ Hein J, Jiang T, Wang L, Zhang K (1996) On the complexity of comparing evolutionary trees. Discrete Applied Mathematics 71:153--169
- ^ Rodrigues, Estela M.; Sagot, Marie-France; Wakabayashi, Yoshiko (April 2007). "The maximum agreement forest problem: Approximation algorithms and computational experiments". Theoretical Computer Science. 374 (1–3): 91–110. doi:10.1016/j.tcs.2006.12.011. S2CID 16479722.
- ^ Kordi M (2019) Inferring Microbial Gene Family Evolution Using Duplication-Transfer-Loss Reconciliation: Algorithms and Complexity. Doctoral Dissertations. 2101.
- ^ Urbini L (2017) Models and algorithms to study the common evolutionary history of hosts and symbionts. Doctoral thesis, Université de Lyon
- ^ Marin, J.; Achaz, G.; Crombach, A.; Lambert, A. (2020). "The genomic view of diversification". Journal of Evolutionary Biology. 33 (10): 1387–1404. doi:10.1111/jeb.13677. PMID 32654283. S2CID 91543153.
- ^ Szöllősi, G. J.; Tannier, E.; Daubin, V.; Boussau, B. (2015). "The inference of gene trees with species trees". Systematic Biology. 64 (1): e42-62. doi:10.1093/sysbio/syu048. PMC 4265139. PMID 25070970.
- ^ Rannala, B.; Yang, Z. (2003). "Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci". Genetics. 164 (4): 1645–1656. doi:10.1093/genetics/164.4.1645. PMC 1462670. PMID 12930768.
- ^ Degnan, J. H.; Salter, L. A. (2005). "Gene tree distributions under the coalescent process". Evolution; International Journal of Organic Evolution. 59 (1): 24–37. doi:10.1111/j.0014-3820.2005.tb00891.x. PMID 15792224.
- ^ Liu, L.; Pearl, D. K. (2007). "Species trees from gene trees: Reconstructing Bayesian posterior distributions of a species phylogeny using estimated gene tree distributions". Systematic Biology. 56 (3): 504–514. doi:10.1080/10635150701429982. PMID 17562474.
- ^ Rannala B and Edwards S and Leaché A and Yang Z (2020) Phylogenetics in the Genomic Era, 3.3:1--3.3:21
- ^ Bork, D.; Cheng, R.; Wang, J.; Sung, J.; Libeskind-Hadas, R. (2017). "On the computational complexity of the maximum parsimony reconciliation problem in the duplication-loss-coalescence model". Algorithms for Molecular Biology. 12: 6. doi:10.1186/s13015-017-0098-8. PMC 5349084. PMID 28316640.
- ^ Chan, Y. B.; Ranwez, V.; Scornavacca, C. (2017). "Inferring incomplete lineage sorting, duplications, transfers and losses with reconciliations" (PDF). Journal of Theoretical Biology. 432: 1–13. Bibcode:2017JThBi.432....1C. doi:10.1016/j.jtbi.2017.08.008. PMID 28801222. S2CID 7854827.
- ^ Du P and Ogilvie H A and Nakhleh L (2019) Unifying Gene Duplication, Loss, and Coalescence on Phylogenetic Networks. Bioinformatics Research and Applications. ISBRA 2019. Lecture Notes in Computer Science, vol 11490. Springer, Cham.
- ^ Rasmussen, M. D.; Kellis, M. (2012). "Unified modeling of gene duplication, loss, and coalescence using a locus tree". Genome Research. 22 (4): 755–765. doi:10.1101/gr.123901.111. PMC 3317157. PMID 22271778.
- ^ Wu, Y. C.; Rasmussen, M. D.; Bansal, M. S.; Kellis, M. (2014). "Most parsimonious reconciliation in the presence of gene duplication, loss, and deep coalescence using labeled coalescent trees". Genome Research. 24 (3): 475–486. doi:10.1101/gr.161968.113. PMC 3941112. PMID 24310000.
- ^ This table is intended to serve as illustration to the 3-Level reconciliation section of Menet et al. (2022) Phylogenetic reconciliation. PLoS Comput Biol 18(11): e1010621.doi:10.1371/journal.pcbi.1010621 and can be read along with it. Colors correspond to the different levels (similar but different colors if there are several trees at the same level). The legend gives an example of all these colours and shape codes. Multiple gene lineages can undergo joint events like whole genome duplication (A) or segmental events (B), some events might be more probable than others, like specific horizontal transfers with highway of transfers or hybridization (C). Cophylogenetic patterns can be compared, to see for instance if the common pattern of a host and a symbiont are not just the common pattern of the symbiont and the geography (D). Characters can evolve on reconciled phylogeny, like gene synteny (E), or two levels can be reconciled with the constraint of an upper one (F). Transfers can be upper dependent, which is more likely between two intermediate entities that belong to a same upper one (G). Three levels can be reconciled together, sequentially: the intermediate and the upper before adding the lower, or trying to find a joint most parsimonious scenario for the two reconciliations (H). These multi-level models can also be used to reconstruct an intermediate phylogeny (I).
- ^ Theis, K. R.; Dheilly, N. M.; Klassen, J. L.; Brucker, R. M.; Baines, J. F.; Bosch, T. C.; Cryan, J. F.; Gilbert, S. F.; Goodnight, C. J.; Lloyd, E. A.; Sapp, J.; Vandenkoornhuyse, P.; Zilber-Rosenberg, I.; Rosenberg, E.; Bordenstein, S. R. (2016). "Getting the Hologenome Concept Right: An Eco-Evolutionary Framework for Hosts and Their Microbiomes". mSystems. 1 (2). doi:10.1128/mSystems.00028-16. PMC 5069740. PMID 27822520.
- ^ Margulis, L. S.; Fester, R. (1991). Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis. Cambridge, Mass.: MIT Press. ISBN 9780262132695.
- ^ Bordenstein, S. R.; Theis, K. R. (2015). "Host Biology in Light of the Microbiome: Ten Principles of Holobionts and Hologenomes". PLOS Biology. 13 (8): e1002226. doi:10.1371/journal.pbio.1002226. PMC 4540581. PMID 26284777.
- ^ Rosenberg, E.; Koren, O.; Reshef, L.; Efrony, R.; Zilber-Rosenberg, I. (2007). "The role of microorganisms in coral health, disease and evolution". Nature Reviews. Microbiology. 5 (5): 355–362. doi:10.1038/nrmicro1635. PMID 17384666. S2CID 2967190.
- ^ Zilber-Rosenberg, I.; Rosenberg, E. (2008). "Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution". FEMS Microbiology Reviews. 32 (5): 723–735. doi:10.1111/j.1574-6976.2008.00123.x. PMID 18549407.
- ^ Moran, N. A.; McCutcheon, J. P.; Nakabachi, A. (2008). "Genomics and evolution of heritable bacterial symbionts". Annual Review of Genetics. 42: 165–190. doi:10.1146/annurev.genet.41.110306.130119. PMID 18983256.
- ^ López-Madrigal, S.; Gil, R. (2017). "Et tu, Brute? Not Even Intracellular Mutualistic Symbionts Escape Horizontal Gene Transfer". Genes. 8 (10): 247. doi:10.3390/genes8100247. PMC 5664097. PMID 28961177.
- ^ Penz, T.; Schmitz-Esser, S.; Kelly, S. E.; Cass, B. N.; Müller, A.; Woyke, T.; Malfatti, S. A.; Hunter, M. S.; Horn, M. (2012). "Comparative genomics suggests an independent origin of cytoplasmic incompatibility in Cardinium hertigii". PLOS Genetics. 8 (10): e1003012. doi:10.1371/journal.pgen.1003012. PMC 3486910. PMID 23133394.
- ^ Nikoh, N.; Hosokawa, T.; Moriyama, M.; Oshima, K.; Hattori, M.; Fukatsu, T. (2014). "Evolutionary origin of insect-Wolbachia nutritional mutualism". Proceedings of the National Academy of Sciences of the United States of America. 111 (28): 10257–10262. Bibcode:2014PNAS..11110257N. doi:10.1073/pnas.1409284111. PMC 4104916. PMID 24982177.
- ^ Manzano-Marı n, A.; Coeur d'Acier, A.; Clamens, A. L.; Orvain, C.; Cruaud, C.; Barbe, V.; Jousselin, E. (2020). "Serial horizontal transfer of vitamin-biosynthetic genes enables the establishment of new nutritional symbionts in aphids' di-symbiotic systems". The ISME Journal. 14 (1): 259–273. Bibcode:2020ISMEJ..14..259M. doi:10.1038/s41396-019-0533-6. PMC 6908640. PMID 31624345.
- ^ Nakabachi, A.; Ueoka, R.; Oshima, K.; Teta, R.; Mangoni, A.; Gurgui, M.; Oldham, N. J.; Van Echten-Deckert, G.; Okamura, K.; Yamamoto, K.; Inoue, H.; Ohkuma, M.; Hongoh, Y.; Miyagishima, S. Y.; Hattori, M.; Piel, J.; Fukatsu, T. (2013). "Defensive bacteriome symbiont with a drastically reduced genome". Current Biology. 23 (15): 1478–1484. Bibcode:2013CBio...23.1478N. doi:10.1016/j.cub.2013.06.027. PMID 23850282. S2CID 1637956.
- ^ Pinto-Carbó, M.; Sieber, S.; Dessein, S.; Wicker, T.; Verstraete, B.; Gademann, K.; Eberl, L.; Carlier, A. (2016). "Evidence of horizontal gene transfer between obligate leaf nodule symbionts". The ISME Journal. 10 (9): 2092–2105. Bibcode:2016ISMEJ..10.2092P. doi:10.1038/ismej.2016.27. PMC 4989318. PMID 26978165.
- ^ Jeong, H.; Arif, B.; Caetano-Anollés, G.; Kim, K. M.; Nasir, A. (2019). "Horizontal gene transfer in human-associated microorganisms inferred by phylogenetic reconstruction and reconciliation". Scientific Reports. 9 (1): 5953. Bibcode:2019NatSR...9.5953J. doi:10.1038/s41598-019-42227-5. PMC 6459891. PMID 30976019.
- ^ Wijayawardena, B. K.; Minchella, D. J.; Dewoody, J. A. (2013). "Hosts, parasites, and horizontal gene transfer". Trends in Parasitology. 29 (7): 329–338. doi:10.1016/j.pt.2013.05.001. PMID 23759418.
- ^ Moodley, Y.; Linz, B.; Bond, R. P.; Nieuwoudt, M.; Soodyall, H.; Schlebusch, C. M.; Bernhöft, S.; Hale, J.; Suerbaum, S.; Mugisha, L.; Van Der Merwe, S. W.; Achtman, M. (2012). "Age of the association between Helicobacter pylori and man". PLOS Pathogens. 8 (5): e1002693. doi:10.1371/journal.ppat.1002693. PMC 3349757. PMID 22589724.
- ^ Achtman, M. (2016). "How old are bacterial pathogens?". Proceedings. Biological Sciences. 283 (1836). doi:10.1098/rspb.2016.0990. PMC 5013766. PMID 27534956.
- ^ Fu, Yiran; Pistolozzi, Marco; Yang, Xiaofeng; Lin, Zhanglin (2020). "A Comprehensive Classification of Coronaviruses and Inferred Cross-Host Transmissions". bioRxiv 10.1101/2020.08.11.232520.
- ^ Da Silva, S. G.; Tehrani, J. J. (2016). "Comparative phylogenetic analyses uncover the ancient roots of Indo-European folktales". Royal Society Open Science. 3 (1): 150645. Bibcode:2016RSOS....350645D. doi:10.1098/rsos.150645. PMC 4736946. PMID 26909191.
- ^ Ross, R. M.; Greenhill, S. J.; Atkinson, Q. D. (2013). "Population structure and cultural geography of a folktale in Europe". Proceedings. Biological Sciences. 280 (1756). doi:10.1098/rspb.2012.3065. PMC 3574383. PMID 23390109.
- ^ Bortolini, E.; Pagani, L.; Crema, E. R.; Sarno, S.; Barbieri, C.; Boattini, A.; Sazzini, M.; Da Silva, S. G.; Martini, G.; Metspalu, M.; Pettener, D.; Luiselli, D.; Tehrani, J. J. (2017). "Inferring patterns of folktale diffusion using genomic data". Proceedings of the National Academy of Sciences of the United States of America. 114 (34): 9140–9145. Bibcode:2017PNAS..114.9140B. doi:10.1073/pnas.1614395114. PMC 5576778. PMID 28784786.
- ^ Guigó, R.; Muchnik, I.; Smith, T. F. (1996). "Reconstruction of ancient molecular phylogeny". Molecular Phylogenetics and Evolution. 6 (2): 189–213. Bibcode:1996MolPE...6..189G. doi:10.1006/mpev.1996.0071. PMID 8899723.
- ^ Page, R. D.; Cotton, J. A. (2002). "Vertebrate phylogenomics: Reconciled trees and gene duplications". Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing: 536–547. doi:10.1142/9789812799623_0050. ISBN 978-981-02-4777-5. PMID 11928506. S2CID 5638166.
- ^ Burleigh, J.G.; Bansal, M.S.; Wehe, A.; Eulenstein, O. (2009). "Locating Large-Scale Gene Duplication Events through Reconciled Trees: Implications for Identifying Ancient Polyploidy Events in Plants". Journal of Computational Biology. 16 (8): 1071–1083. doi:10.1089/cmb.2009.0139. PMID 19689214.
- ^ Bansal, M. S.; Eulenstein, O. (2008). "The multiple gene duplication problem revisited". Bioinformatics. 24 (13): i132-8. doi:10.1093/bioinformatics/btn150. PMC 2718628. PMID 18586705.
- ^ Michael R. Fellows, Michael T. Hallet, and Ulrike Stege. 1998. On the Multiple Gene Duplication Problem. In Proceedings of the 9th International Symposium on Algorithms and Computation (ISAAC '98). Springer-Verlag, Berlin, Heidelberg, 347–356.
- ^ Dondi, R.; Lafond, M.; Scornavacca, C. (2019). "Reconciling multiple genes trees via segmental duplications and losses". Algorithms for Molecular Biology. 14: 7. doi:10.1186/s13015-019-0139-6. PMC 6425616. PMID 30930955.
- ^ Bansal, M. S.; Banay, G.; Gogarten, J. P.; Shamir, R. (2011). "Detecting highways of horizontal gene transfer". Journal of Computational Biology. 18 (9): 1087–1114. doi:10.1089/cmb.2011.0066. hdl:1721.1/69855. PMID 21899418.
- ^ Kloub, L.; Gosselin, S.; Fullmer, M.; Graf, J.; Gogarten, J. P.; Bansal, M. S. (2021). "Systematic Detection of Large-Scale Multigene Horizontal Transfer in Prokaryotes". Molecular Biology and Evolution. 38 (6): 2639–2659. doi:10.1093/molbev/msab043. PMC 8136488. PMID 33565580.
- ^ Yu, Y.; Ristic, N.; Nakhleh, L. (2013). "Fast algorithms and heuristics for phylogenomics under ILS and hybridization". BMC Bioinformatics. 14 (Suppl 15): S6. doi:10.1186/1471-2105-14-S15-S6. PMC 3852049. PMID 24564257.
- ^ Yu, Y.; Barnett, R. M.; Nakhleh, L. (2013). "Parsimonious inference of hybridization in the presence of incomplete lineage sorting". Systematic Biology. 62 (5): 738–751. doi:10.1093/sysbio/syt037. PMC 3739885. PMID 23736104.
- ^ Nieberding, C. M.; Durette-Desset, M. C.; Vanderpoorten, A.; Casanova, J. C.; Ribas, A.; Deffontaine, V.; Feliu, C.; Morand, S.; Libois, R.; Michaux, J. R. (2008). "Geography and host biogeography matter for understanding the phylogeography of a parasite". Molecular Phylogenetics and Evolution. 47 (2): 538–554. Bibcode:2008MolPE..47..538N. doi:10.1016/j.ympev.2008.01.028. hdl:2268/9909. PMID 18346916.
- ^ Martínez-Aquino, A.; Ceccarelli, F. S.; Eguiarte, L. E.; Vázquez-Domínguez, E.; De León, G. P. (2014). "Do the historical biogeography and evolutionary history of the digenean Margotrema SPP. Across central Mexico mirror those of their freshwater fish hosts (Goodeinae)?". PLOS ONE. 9 (7): e101700. Bibcode:2014PLoSO...9j1700M. doi:10.1371/journal.pone.0101700. PMC 4084993. PMID 24999998.
- ^ Weckstein, J. D. (2004). "Biogeography explains cophylogenetic patterns in toucan chewing lice". Systematic Biology. 53 (1): 154–164. doi:10.1080/10635150490265085. PMID 14965910.
- ^ Fountain, E. D.; Pauli, J. N.; Mendoza, J. E.; Carlson, J.; Peery, M. Z. (2017). "Cophylogenetics and biogeography reveal a coevolved relationship between sloths and their symbiont algae". Molecular Phylogenetics and Evolution. 110: 73–80. Bibcode:2017MolPE.110...73F. doi:10.1016/j.ympev.2017.03.003. PMID 28288943.
- ^ Groussin, M.; Mazel, F.; Sanders, J. G.; Smillie, C. S.; Lavergne, S.; Thuiller, W.; Alm, E. J. (2017). "Unraveling the processes shaping mammalian gut microbiomes over evolutionary time". Nature Communications. 8: 14319. Bibcode:2017NatCo...814319G. doi:10.1038/ncomms14319. PMC 5331214. PMID 28230052.
- ^ Berry, V.; Chevenet, F.; Doyon, J. P.; Jousselin, E. (2018). "A geography-aware reconciliation method to investigate diversification patterns in host/Parasite interactions". Molecular Ecology Resources. 18 (5): 1173–1184. doi:10.1111/1755-0998.12897. PMID 29697894. S2CID 25758856.
- ^ Wu, Y. C.; Rasmussen, M. D.; Kellis, M. (2012). "Evolution at the subgene level: Domain rearrangements in the Drosophila phylogeny". Molecular Biology and Evolution. 29 (2): 689–705. doi:10.1093/molbev/msr222. PMC 3258039. PMID 21900599.
- ^ a b Stolzer, M.; Siewert, K.; Lai, H.; Xu, M.; Durand, D. (2015). "Event inference in multidomain families with phylogenetic reconciliation". BMC Bioinformatics. 16 (Suppl 14): S8. doi:10.1186/1471-2105-16-S14-S8. PMC 4610023. PMID 26451642.
- ^ a b Li, L.; Bansal, M. S. (2019). "An Integrated Reconciliation Framework for Domain, Gene, and Species Level Evolution". IEEE/ACM Transactions on Computational Biology and Bioinformatics. 16 (1): 63–76. doi:10.1109/TCBB.2018.2846253. PMID 29994126. S2CID 51614715.
- ^ Li, Lei; Bansal, Mukul S. (2018). "An Integer Linear Programming Solution for the Domain-Gene-Species Reconciliation Problem". Proceedings of the 2018 ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics. pp. 386–397. doi:10.1145/3233547.3233603. ISBN 9781450357944. S2CID 49426403.
- ^ Li, Lei; Bansal, Mukul S. (2019). "Simultaneous Multi-Domain-Multi-Gene Reconciliation Under the Domain-Gene-Species Reconciliation Model". Bioinformatics Research and Applications. Lecture Notes in Computer Science. Vol. 11490. pp. 73–86. doi:10.1007/978-3-030-20242-2_7. ISBN 978-3-030-20241-5. S2CID 85507475.
- ^ Kundu, S.; Bansal, M. S. (2019). "SaGePhy: An improved phylogenetic simulation framework for gene and subgene evolution". Bioinformatics. 35 (18): 3496–3498. doi:10.1093/bioinformatics/btz081. PMID 30715213.
- ^ Muhammad, Sayyed Auwn; Sennblad, Bengt; Lagergren, Jens (2018). "Species tree-aware simultaneous reconstruction of gene and domain evolution". doi:10.1101/336453. S2CID 90627553.
{{cite journal}}: Cite journal requires|journal=(help) - ^


 French
French Deutsch
Deutsch