ShelXle
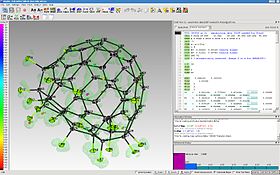 the ShelXle main window with a structure of C60F18 | |
| Developer(s) | Christian B. Hübschle |
|---|---|
| Initial release | 2011 |
| Stable release | 1.0.1229[1] |
| Written in | C++ |
| Operating system | Windows, Linux, macOS |
| Type | Molecular modelling |
| License | GNU Lesser General Public License |
| Website | http://www.shelxle.org/index.php |
The program ShelXle[2] is a graphical user interface for the structure refinement program SHELXL.[3] ShelXle combines an editor with syntax highlighting for the SHELXL-associated .ins (input) and .res (output) files with an interactive graphical display for visualization of a three-dimensional structure including the electron density (Fo) and difference density (Fo-Fc) maps.
Overview
[edit]ShelXle can display electron density maps like the macromolecular program Coot but is more intended for smaller molecules. A number of excellent graphical user interfaces (GUIs) exist for small molecule crystal structure refinement with SHELX (e.g., WINGX,[4] Olex2,[5] XSEED,[6] PLATON and SYSTEM-S,[7] and the Bruker programs XP and XSHELL)
ShelXle is free software, distributed under the GNU LGPL. It is available from the ShelXle website or from SourceForge.[8][9] Binaries are available for Windows, macOS and the Linux distributions SuSE, Debian and Ubuntu. The Windows binary is distributed with the NSIS Installer.
Features
[edit]- Editor featuring syntax highlighting and code completion for the SHELX instructions.
- Clicking on an atom in the structure view sets the text cursor to the line that contains this atom.
- Locating atoms in structure view from the editor.
- Rename mode with support of residues and disordered parts and free variables.
Program architecture
[edit]ShelXle uses the Qt (framework). It is written entirely in C++ and does not use any scripting language. For the refinement it calls the external binary of SHELXL which might also be SHELXH, SHELXLMP from George M. Sheldrick or XL from Bruker.
SHELX
[edit]SHELX[3] is developed by George M. Sheldrick since the late 1960s. Important releases are SHELX76 and SHELX97. It is still developed but releases are usually after ten years of testing. Academic users can download the SHELX programs freely after registration.[10]
External links
[edit]References
[edit]- ^ https://www.shelxle.org/shelx/chlg/changelog.php.
{{cite web}}: Missing or empty|title=(help) - ^ Hübschle, Christian B.; Sheldrick, George M.; Dittrich, Birger (12 November 2011). "ShelXle: a Qt graphical user interface for SHELXL". Journal of Applied Crystallography. 44 (6): 1281–1284. doi:10.1107/S0021889811043202. PMC 3246833. PMID 22477785.
- ^ a b Sheldrick, George M. (21 December 2007). "A short history of SHELX". Acta Crystallographica Section A. 64 (1): 112–122. Bibcode:2008AcCrA..64..112S. doi:10.1107/S0108767307043930. PMID 18156677.
- ^ Farrugia, Louis J. (1 August 1999). "WinGX suite for small-molecule single-crystal crystallography". Journal of Applied Crystallography. 32 (4): 837–838. doi:10.1107/S0021889899006020.
- ^ Dolomanov, Oleg V.; Bourhis, Luc J.; Gildea, Richard J.; Howard, Judith A. K.; Puschmann, Horst (24 January 2009). "OLEX2: a complete structure solution, refinement and analysis program". Journal of Applied Crystallography. 42 (2): 339–341. doi:10.1107/S0021889808042726.
- ^ Barbour, Leonard J. (2001). "X-Seed — A Software Tool for Supramolecular Crystallography". J. Supramol. Chem. 1 (4–6): 189–19. doi:10.1016/S1472-7862(02)00030-8.
- ^ Spek, Anthony L. (20 January 2009). "Structure validation in chemical crystallography". Acta Crystallographica Section D. 65 (2): 148–155. doi:10.1107/S090744490804362X. PMC 2631630. PMID 19171970.
- ^ "ShelXle Download Page". www.shelxle.org. Retrieved 2021-09-15.
- ^ "ShelXle". SourceForge. Retrieved 2021-09-15.
- ^ "The SHELX homepage". shelx.uni-goettingen.de. Retrieved 2016-09-06.


 French
French Deutsch
Deutsch