Substructure search

Substructure search (SSS) is a method to retrieve from a database only those chemicals matching a pattern of atoms and bonds which a user specifies. It is an application of graph theory, specifically subgraph matching in which the query is a hydrogen-depleted molecular graph. The mathematical foundations for the method were laid in the 1870s, when it was suggested that chemical structure drawings were equivalent to graphs with atoms as vertices and bonds as edges. SSS is now a standard part of cheminformatics and is widely used by pharmaceutical chemists in drug discovery.
There are many commercial systems that provide SSS, typically having a graphical user interface and chemical drawing software. Large publicly-available databases like PubChem and ChemSpider can be searched this way, as can Wikipedia's articles describing individual chemicals.
Definitions
[edit]Substructure search is used to retrieve from a database of chemicals those which contain the pattern of atoms and bonds specified by a user. It is implemented using a specialist type of query language and in real-world applications the search may be further constrained using logical operators on additional data held in the database. Thus "return all carboxylic acids where a sample of >1 g is available".[1][2] One definition of "substructure" was provided in 2008: "given two chemical structures A and B, if structure A is fully contained in structure B, then A is a substructure of B, while B is a superstructure of A."[3]
molecular graph: The graph with differently labelled (coloured) vertices (chromatic graph) which represent different kinds of atoms and differently labelled (coloured) edges related to different types of bonds. Within the topological electron distribution theory, a complete network of the bond paths for a given nuclear configuration.[4]
In this definition, the word "structure" is not synonymous with "compound". If it were, the structure for ethanol, CH3CH2OH would not be a substructure of propanol, CH3CH2CH2OH, since the terminal CH3 of ethanol is not fully contained at the propanol chain two atoms away from the OH group. Instead the query structure is, formally, a hydrogen-depleted molecular graph. The search is thus for substances which contain three atoms and two single bonds connected as C–C–O. Propanol is a "hit", as is diethyl ether, with C–C–O–C–C. If a user wished to limit the hits to alcohols, then the query structure would have to be drawn with an "explicit hydrogen", as C–C–O–H and ether would no longer match.[1] In mathematical terms, finding substructures is an application of graph theory, specifically subgraph matching.[5]
Examples
[edit]Standard conventions used when chemists draw chemical structures[6] need to be considered when implementing substructure search. Historically, the representation of tautomer[7] forms and stereochemistry[8] has posed difficulties. This can be illustrated using histidine.[9]
The top row shows the standard two-dimensional chemical drawing for (S)-histidine (the natural isomer of this amino acid), its enantiomer (R)-histidine and a drawing which conventionally indicates the racemic mixture of equal amounts of the R and S forms.[10] The bottom row shows the same three compounds with the imidazole ring drawn in its alternative tautomer form. For histidine, it has been experimentally determined by 15N NMR spectroscopy that the 1-H tautomer is preferred over the 3-H form in samples.[11] Choice of representation for storage in a database can influence substucture searches. All six drawings are hits for a propanol substructure C–C–C–O, as shown in red. However, only the top row would, apparently, be a hit for the blue substructure of 1-H imidazole-4-methyl, as this is not fully contained in the other three compounds. In fact, each vertical pair is the same chemical substance: tautomers in general cannot be isolated as separate samples.[7] In modern databases, substances are held in a single canonical form, with checks made for uniqueness. The InChIKey provides one way to do this.[9] (S)-Histidine's standard key is HNDVDQJCIGZPNO-YFKPBYRVSA-N,[12] (R)-histidine's key is HNDVDQJCIGZPNO-RXMQYKEDSA-N[13] and (RS)-histidine's is HNDVDQJCIGZPNO-UHFFFAOYSA-N.[14] The first block of 14 letters is identical for all these substances, as it encodes the molecular graph.[9]
Query interfaces and search algorithms
[edit]Most substructure search systems present the user with a graphical user interface with a chemical structure drawing component. Query structures may contain bonding patterns such as "single/aromatic" or "any" to provide flexibility. Similarly, the vertices which in an actual compound would be a specific atom may be replaced with an atom list in the query. Cis–trans isomerism at double bonds is catered for by giving a choice of retrieving only the E form, the Z form, or both.[1][15]
The algorithms for searching are computationally intensive, often of O (n3) or O (n4) time complexity (where n is the number of atoms involved) but the problem is known to be NP-complete.[16] Speedups are achieved using fragment screening as a first step. This pre-computation typically involves creation of bitstrings representing presence or absence of molecular fragments. Target compounds that do not possess the fragments present in the query cannot be hits and are eliminated.[17][18] Atom-by-atom-searching, in which a mapping of the query's atoms and bonds with the target molecule is sought, is usually done with a variant of the Ullman algorithm.[5][19]
Implementations
[edit]As of 2024[update], substructure search is a standard feature in chemical databases accessible via the web. Large databases such as PubChem,[20][15] maintained by the National Center for Biotechnology Information and ChemSpider,[21] maintained by the Royal Society of Chemistry have graphical interfaces for search. The Chemical Abstracts Service, a division of the American Chemical Society, provides tools to search the chemical literature and Reaxys supplied by Elsevier covers both chemicals and reaction information, including that originally held in the Beilstein database.[22] PATENTSCOPE maintained by the World Intellectual Property Organization makes chemical patents accessible by substructure[23] and Wikipedia's articles describing individual chemicals can also be searched that way.[24]
Suppliers of chemicals as synthesis intermediates or for high-throughput screening routinely provide search interfaces. Currently, the largest database that can be freely searched by the public is the ZINC database, which is claimed to contain over 37 billion commercially available molecules.[25][26]
History
[edit]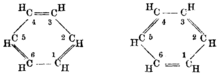
The idea that chemical structures as depicted using drawings of the type introduced by Kekulé were related to what is now called graph theory was suggested by the mathematician J. J. Sylvester in 1878. He was the first to use the word "graph" in the sense of a network.[27][28] Arthur Cayley had already, in 1874, considered how to enumerate chemical isomers, in what was an early approach to molecular graphs, where atoms are at vertices and bonds correspond to edges.[29][30]
structural formula: A formula which gives information about the way the atoms in a molecule are connected and arranged in space.[31]
In the 20th century, chemists developed standard ways to show structural formula, especially for individual organic compounds that were increasingly being synthesized and tested as potential drugs or agrochemicals,[32][6] By the 1950s, as the number of compounds made and tested grew, the first attempts to create chemical databases were made and the sub-discipline of cheminformatics was established.[33] As stated in 2012, "searching for substructures in molecules belongs to the most elementary tasks in cheminformatics and is nowadays part of virtually every cheminformatics software".[34]
The first suggested use for substructure search was in 1957, to reduce the workload of patent examiners. They have to search published literature to decide whether an invention is novel, which for chemical patents often means finding known examples within the generic claims of a Markush structure.[35][33] Before this could become a reality, a number of developments were required. Importantly, the existing literature had to be made searchable and a way to input a chemical structure query and return the matching results had to devised. These requirements had been partially met as early as 1881 when Friedrich Konrad Beilstein introduced the Handbuch der organischen Chemie (Handbook of Organic Chemistry) which carefully classified known chemicals in a very systematic manner so that all examples containing a given heterocycle would be located together.[36][37]
In 1907, the American Chemical Society set up the Chemical Abstracts Service (CAS). This weekly subscription service included a printed publication with summaries of articles in thousands of scholarly journals and claims in worldwide patents. This had a chemical substance index that, in principle, allowed searching by chemical name or formula.[38] However, it was only when the CAS records had been fully converted into machine-readable form and the internet was available to connect its database to end-users that comprehensive searching became possible. CAS provided various specialist search services from the 1980s but it was not until 2008 that its "SciFinder" system became available via the web.[39]
By the 1960s, companies synthesizing and testing new chemicals made significant progress in creating in-house databases. Imperial Chemical Industries stored chemical structures encoded as text strings, using Wiswesser line notation. Its associated CROSSBOW software allowed substructure search using key-based searches followed by more processor-intensive atom-by-atom search.[40][41] It was recognised that research chemists wanted not only to search company collections for existing inventory but also to search third-party databases supplied by vendors of small-molecule intermediates. The latter application evolved as a collaboration involving six companies with pharmaceutical interests and their commercial suppliers.[42][9]
By the 1980s, other line notations were used for commercially-available substructure search systems. SMILES encoding, together with its SMARTS query language,[43] and SYBYL line notation[9][44] are examples.[45] A comprehensive survey of then-available chemical information systems was produced for NASA in 1985.[46]
The need to combine chemistry search with biological data produced by screening compounds at ever-larger scales led to implementation of systems such as MACCS.[46]: 73–77 [47] This commercial system from MDL Information Systems made use of an algorithm specifically designed for storage and search within groups of chemicals that differed only in their stereochemistry.[48] A review of the many systems available by the mid-1980s pointed out that "most in-house developed systems have been replaced with commercially available standardised software for managing chemical structure databases."[49] The MDL Molfile is now an open file format for storing single-molecule data in the form of a connection table.[50][9]
By the 2000s, personal computers had become powerful enough that storage and search of chemistry within office software such as Microsoft Excel was possible.[51]
Subsequent developments involved the use of new techniques to allow efficient searches over very large databases and, importantly, the use of a standardised International Chemical Identifier, a type of line notation, to uniquely define a chemical substance.[9][25][52][53]
See also
[edit]References
[edit]- ^ a b c Currano, Judith N. (2014). "Chapter 5. Searching by Structure and Substructure". Chemical Information for Chemists. pp. 109–145. doi:10.1039/9781782620655-00109. ISBN 978-1-84973-551-3.
- ^ Agrafiotis, Dimitris K.; Lobanov, Victor S.; Shemanarev, Maxim; et al. (2011). "Efficient Substructure Searching of Large Chemical Libraries: The ABCD Chemical Cartridge". Journal of Chemical Information and Modeling. 51 (12): 3113–3130. doi:10.1021/ci200413e. PMID 22035187.
- ^ Cao, Yiqun; Jiang, Tao; Girke, Thomas (2008). "A maximum common substructure-based algorithm for searching and predicting drug-like compounds". Bioinformatics. 24 (13): i366 – i374. doi:10.1093/bioinformatics/btn186. PMC 2718661. PMID 18586736.
- ^ "molecular graph". Gold Book. IUPAC. 2014. doi:10.1351/goldbook.MT07069.
- ^ a b Ullmann, J. R. (1976). "An Algorithm for Subgraph Isomorphism". Journal of the ACM. 23: 31–42. doi:10.1145/321921.321925.
- ^ a b McMurry, John (2023). "1.12 Drawing Chemical Structures". Organic Chemistry: A Tenth Edition. OpenStax, Rice University. pp. 25–27. ISBN 9781711471853.
- ^ a b Katritzky, Alan R.; Hall, C. Dennis; El-Gendy, Bahaa El-Dien M.; Draghici, Bogdan (2010). "Tautomerism in drug discovery". Journal of Computer-Aided Molecular Design. 24 (6–7): 475–484. Bibcode:2010JCAMD..24..475K. doi:10.1007/s10822-010-9359-z. PMID 20490619.
- ^ Smith, Silas W. (2009). "Chiral Toxicology: It's the Same Thing…Only Different". Toxicological Sciences. 110 (1): 4–30. doi:10.1093/toxsci/kfp097. PMID 19414517.
- ^ a b c d e f g Warr, Wendy A. (2011). "Representation of chemical structures". WIREs Computational Molecular Science. 1 (4): 557–579. doi:10.1002/wcms.36.
- ^ "Search term: histidine". chemspider.com. Retrieved 2024-08-01.
- ^ Roberts, John D. (2000). ABCs of FT-NMR. Sausalito, CA: University Science Books. pp. 258–9. ISBN 978-1-891389-18-4.
- ^ "L-Histidine". chemspider.com. More details. Retrieved 2024-08-01.
- ^ "D-Histidine". chemspider.com. More details. Retrieved 2024-08-01.
- ^ "DL-Histidine". chemspider.com. More details. Retrieved 2024-08-01.
- ^ a b "PubChem Structure Search". pubchem.ncbi.nlm.nih.gov. Retrieved 2024-08-01.
- ^ Wegener, Ingo (2005). Complexity Theory: Exploring the Limits of Efficient Algorithms. Springer. p. 81. ISBN 9783540210450.
- ^ Bond, V. Lynn; Bowman, Carlos M.; Davison, Linda C.; et al. (1979). "On-Line Storage and Retrieval of Chemical Information. II. Substructure and Biological Activity Searching". Journal of Chemical Information and Computer Sciences. 19 (4): 231–234. doi:10.1021/ci60020a012. PMID 551973.
- ^ Cummings, Maxwell D.; Maxwell, Alan C.; DesJarlais, Renee L. (2007). "Processing of Small Molecule Databases for Automated Docking". Medicinal Chemistry. 3 (1): 107–113. doi:10.2174/157340607779317481. PMID 17266630.
- ^ Rahman, S. A.; Bashton, M.; Holliday, G. L.; Schrader, R.; Thornton, J. M. (2000). "Small Molecule Subgraph Detector (SMSD) toolkit". Journal of Cheminformatics. 1 (1): 12. doi:10.1186/1758-2946-1-12. PMC 2820491. PMID 20298518.
- ^ Kim, Sunghwan (2021). "Exploring Chemical Information in PubChem". Current Protocols. 1 (8): e217. doi:10.1002/cpz1.217. PMC 8363119. PMID 34370395.
- ^ Williams, Antony J. (2010). "ChemSpider: Integrating Structure-Based Resources Distributed across the Internet". Enhancing Learning with Online Resources, Social Networking, and Digital Libraries. ACS Symposium Series. Vol. 1060. pp. 23–39. doi:10.1021/bk-2010-1060.ch002. ISBN 978-0-8412-2600-5.
- ^ Jarabak, Charlotte; Mutton, Troy; Ridley, Damon D. (2020). "Property Information in Substance Records in Major Web-Based Chemical Information and Data Retrieval Tools: Understanding Content, Search Opportunities, and Application to Teaching". Journal of Chemical Education. 97 (5): 1345–1359. Bibcode:2020JChEd..97.1345J. doi:10.1021/acs.jchemed.9b00966.
- ^ "Substructure Search Now Available in PATENTSCOPE". www.wipo.int. 2019-02-11. Retrieved 2024-08-04.
- ^ Ertl, Peter; Patiny, Luc; Sander, Thomas; et al. (2015). "Wikipedia Chemical Structure Explorer: Substructure and similarity searching of molecules from Wikipedia". Journal of Cheminformatics. 7: 10. doi:10.1186/s13321-015-0061-y. PMC 4374119. PMID 25815062.
- ^ a b Tingle, Benjamin I.; Tang, Khanh G.; Castanon, Mar; Gutierrez, John J.; Khurelbaatar, Munkhzul; Dandarchuluun, Chinzorig; Moroz, Yurii S.; Irwin, John J. (2023). "ZINC-22─A Free Multi-Billion-Scale Database of Tangible Compounds for Ligand Discovery". Journal of Chemical Information and Modeling. 63 (4): 1166–1176. doi:10.1021/acs.jcim.2c01253. PMC 9976280. PMID 36790087.
- ^ Warr, Wendy A.; Nicklaus, Marc C.; Nicolaou, Christos A.; Rarey, Matthias (2022). "Exploration of Ultralarge Compound Collections for Drug Discovery". Journal of Chemical Information and Modeling. 62 (9): 2021–2034. doi:10.1021/acs.jcim.2c00224. PMID 35421301.
- ^ Sylvester, J. J. (1878). "Chemistry and Algebra". Nature. 17 (432): 284. Bibcode:1878Natur..17..284S. doi:10.1038/017284a0.
Every invariant and covariant thus becomes expressible by a graph precisely identical with a Kekuléan diagram or chemicograph.
- ^ Gross, Jonathan L.; Yellen, Jay (2004). Handbook of graph theory. CRC Press. p. 35. ISBN 978-1-58488-090-5. Retrieved 2024-07-28.
- ^ Cayley (1874). "LVII. On the mathematical theory of isomers". The London, Edinburgh, and Dublin Philosophical Magazine and Journal of Science. 47 (314): 444–447. doi:10.1080/14786447408641058.
- ^ Biggs, Norman; Keith Lloyd, E.; Wilson, Robin J. (1986). Graph Theory, 1736-1936. Clarendon Press. pp. 39, 63–64. ISBN 0198539169.
- ^ "structural formula". Gold Book. IUPAC. 2014. doi:10.1351/goldbook.S06061.
- ^ Goodwin, W. M. (2008). "Structural formulas and explanation in organic chemistry". Foundations of Chemistry. 10 (2): 117–127. doi:10.1007/s10698-007-9033-2.
- ^ a b Willett, Peter (2008). "From chemical documentation to chemoinformatics: 50 years of chemical information science". Journal of Information Science. 34 (4): 477–499. doi:10.1177/0165551507084631.
- ^ Ehrlich, Hans-Christian; Rarey, Matthias (2012). "Systematic benchmark of substructure search in molecular graphs - from Ullmann to VF2". Journal of Cheminformatics. 4 (1): 13. doi:10.1186/1758-2946-4-13. PMC 3586954. PMID 22849361.
- ^ Ray, Louis C.; Kirsch, Russell A. (1957). "Finding Chemical Records by Digital Computers". Science. 126 (3278): 814–819. Bibcode:1957Sci...126..814R. doi:10.1126/science.126.3278.814. PMID 17776535.
- ^ Richter, Friedrich (1938). "How Beilstein is made". Journal of Chemical Education. 15 (7): 310. Bibcode:1938JChEd..15..310R. doi:10.1021/ed015p310.
- ^ White, Michael J. (2014). "Chapter 3. Chemical Patents". Chemical Information for Chemists. pp. 53–90. doi:10.1039/9781782620655-00053. ISBN 978-1-84973-551-3.
- ^ "CAS Printed Products". CAS. Archived from the original on 2008-05-12. Retrieved 2024-07-29.
- ^ "New SciFinder Available Via the Web". CAS. Archived from the original on 2008-05-13. Retrieved 2024-07-29.
- ^ Eakin, Diane R.; Hyde, Ernest; Palmer, Graham (1974). "The use of computers with chemical structural information: ICI CROSSBOW system". Pesticide Science. 5 (3): 319–326. doi:10.1002/ps.2780050316.
- ^ Warr, Wendy A. (1982). "Diverse uses and future prospects for Wiswesser line-formula notation". Journal of Chemical Information and Computer Sciences. 22 (2): 98–101. doi:10.1021/ci00034a007.
- ^ Walker, S. Barrie (1983). "Development of CAOCI and its use in ICI plant protection division". Journal of Chemical Information and Computer Sciences. 23: 3–5. doi:10.1021/ci00037a001.
- ^ Weininger, David (1988). "SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules". Journal of Chemical Information and Computer Sciences. 28: 31–36. doi:10.1021/ci00057a005.
- ^ Homer, R. Webster; Swanson, Jon; Jilek, Robert J.; et al. (2008). "SYBYL Line Notation (SLN): A Single Notation to Represent Chemical Structures, Queries, Reactions, and Virtual Libraries". Journal of Chemical Information and Modeling. 48 (12): 2294–2307. doi:10.1021/ci7004687. PMID 18998666.
- ^ Wiswesser, William J. (1985). "Historic development of chemical notations". Journal of Chemical Information and Computer Sciences. 25 (3): 258–263. doi:10.1021/ci00047a023.
- ^ a b Shaik, Aneesa Bashir (1985-12-05). "A survey of chemical information systems" (PDF). ntrs.nasa.gov. pp. 1–160.
- ^ Adamson, George W.; Bird, John M.; Palmer, Graham; Warr, Wendy A. (1985). "Use of MACCS within ICI". Journal of Chemical Information and Computer Sciences. 25 (2): 90–92. doi:10.1021/ci00046a007.
- ^ Wipke, W. Todd; Dyott, Thomas M. (1974). "Stereochemically unique naming algorithm". Journal of the American Chemical Society. 96 (15): 4834–4842. Bibcode:1974JAChS..96.4834W. doi:10.1021/ja00822a021.
- ^ Hagadone, Tom R. (1988). "Current Approaches and New Directions in the Management of In-House Chemical Structure Databases". Chemical Structures. pp. 23–41. doi:10.1007/978-3-642-73975-0_3. ISBN 978-3-642-73977-4.
- ^ "CT File Formats" (PDF). Biovia. August 2020. Archived (PDF) from the original on 2021-02-19. Retrieved 2024-08-01.
- ^ Lawson, Kevin R.; Lawson, Jonty (2012). "LICSS - a chemical spreadsheet in microsoft excel". Journal of Cheminformatics. 4 (1): 3. doi:10.1186/1758-2946-4-3. PMC 3310842. PMID 22301088.
- ^ Judson, Philip (2019). "Chapter 7. Structure, Substructure and Superstructure Searching". Knowledge-based Expert Systems in Chemistry. Theoretical and Computational Chemistry Series. Royal Society of Chemistry. pp. 84–107. doi:10.1039/9781788016186-00084. ISBN 978-1-78801-471-7.
- ^ Rarey, Matthias; Nicklaus, Marc C.; Warr, Wendy (2022). "Special Issue on Reaction Informatics and Chemical Space". Journal of Chemical Information and Modeling. 62 (9): 2009–2010. doi:10.1021/acs.jcim.2c00390. PMID 35527682.
External links
[edit]- Wikipedia Chemical Structure Explorer to search Wikipedia chemistry articles by substructure
- Search PubChem
- Search ChemSpider
- Search ZINC-22, a database of over 50 billion molecules


 French
French Deutsch
Deutsch