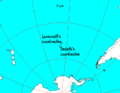
print "import modules...", import sys sys.stdout.flush() import pickle from mpl_toolkits.basemap import Basemap, shiftgrid, cm import matplotlib import matplotlib.pyplot as plt import numpy as np from netCDF4 import Dataset print "ok" # Lovecraft: 47:9'S 126:43'W lovecraft_lat = -47.9 lovecraft_lon = -126.43 # August Derleth: 49:51'S 128:34'W derleth_lat = -49.51 derleth_lon = -128.34 # Nemo point: 48:52.6'S 123:23.6'W nemo_lat = -48.526 nemo_lon = -123.236 # The Bloop: # Appears to be way too far from the Nemo point to be interesting in a R'lyeh context # bransfield_strait_lat=-63 # bransfield_strait_lon=-59 # ross_sea_lat = -75 # ross_sea_lon = -175 # cape_adare_lat = -71.17 # cape_adare_lon = -170.14 # Not necessary, because the default projection is ortho, # but can be useful if you want another one. def equi(m, centerlon, centerlat, radius, *args, **kwargs): """ Drawing circles of a given radius around any point on earth, given the current projection. http://www.geophysique.be/2011/02/20/matplotlib-basemap-tutorial-09-drawing-circles/ """ glon1 = centerlon glat1 = centerlat X = [] Y = [] for azimuth in range(0, 360): glon2, glat2, baz = shoot(glon1, glat1, azimuth, radius) X.append(glon2) Y.append(glat2) X.append(X[0]) Y.append(Y[0]) #m.plot(X,Y,**kwargs) #Should work, but doesn't... X,Y = m(X,Y) plt.plot(X,Y,**kwargs) def shoot(lon, lat, azimuth, maxdist=None): """Shooter Function Plotting great circles with Basemap, but knowing only the longitude, latitude, the azimuth and a distance. Only the origin point is known. Original javascript on http://williams.best.vwh.net/gccalc.htm Translated to python by Thomas Lecocq : http://www.geophysique.be/2011/02/19/matplotlib-basemap-tutorial-08-shooting-great-circles/ """ glat1 = lat * np.pi / 180. glon1 = lon * np.pi / 180. s = maxdist / 1.852 faz = azimuth * np.pi / 180. EPS= 0.00000000005 if ((np.abs(np.cos(glat1))<EPS) and not (np.abs(np.sin(faz))<EPS)): alert("Only N-S courses are meaningful, starting at a pole!") a=6378.13/1.852 f=1/298.257223563 r = 1 - f tu = r * np.tan(glat1) sf = np.sin(faz) cf = np.cos(faz) if (cf==0): b=0. else: b=2. * np.arctan2 (tu, cf) cu = 1. / np.sqrt(1 + tu * tu) su = tu * cu sa = cu * sf c2a = 1 - sa * sa x = 1. + np.sqrt(1. + c2a * (1. / (r * r) - 1.)) x = (x - 2.) / x c = 1. - x c = (x * x / 4. + 1.) / c d = (0.375 * x * x - 1.) * x tu = s / (r * a * c) y = tu c = y + 1 while (np.abs (y - c) > EPS): sy = np.sin(y) cy = np.cos(y) cz = np.cos(b + y) e = 2. * cz * cz - 1. c = y x = e * cy y = e + e - 1. y = (((sy * sy * 4. - 3.) * y * cz * d / 6. + x) * d / 4. - cz) * sy * d + tu b = cu * cy * cf - su * sy c = r * np.sqrt(sa * sa + b * b) d = su * cy + cu * sy * cf glat2 = (np.arctan2(d, c) + np.pi) % (2*np.pi) - np.pi c = cu * cy - su * sy * cf x = np.arctan2(sy * sf, c) c = ((-3. * c2a + 4.) * f + 4.) * c2a * f / 16. d = ((e * cy * c + cz) * sy * c + y) * sa glon2 = ((glon1 + x - (1. - c) * d * f + np.pi) % (2*np.pi)) - np.pi baz = (np.arctan2(sa, b) + np.pi) % (2 * np.pi) glon2 *= 180./np.pi glat2 *= 180./np.pi baz *= 180./np.pi return (glon2, glat2, baz) print "read in etopo5 topography/bathymetry" url = 'http://ferret.pmel.noaa.gov/thredds/dodsC/data/PMEL/etopo5.nc' etopodata = Dataset(url) print "get data" def topopickle(etopodata,name): import sys print "\t"+name+"...", sys.stdout.flush() filename = "rlyeh_"+name+".pickle" try: with open(filename,"r") as fd: print "load...", var = pickle.load(fd) except IOError: print "copy...", var = etopodata.variables[name][:] with open(filename,"w") as fd: print "dump...", pickle.dump(var,fd) print "ok" return var topoin = topopickle(etopodata,"ROSE") lons = topopickle(etopodata,"ETOPO05_X") lats = topopickle(etopodata,"ETOPO05_Y") print "shift data so lons go from -180 to 180 instead of 20 to 380...", sys.stdout.flush() topoin,lons = shiftgrid(180.,topoin,lons,start=False) print "ok" # create the figure and axes instances. fig = plt.figure() ax = fig.add_axes([0.1,0.1,0.8,0.8]) print "setup basemap" # set up orthographic m projection with # perspective of satellite looking down at 50N, 100W. # use low resolution coastlines. m = Basemap(projection='ortho',lat_0=nemo_lat,lon_0=nemo_lon,resolution='l') m.bluemarble() # Generic Mapping Tools colormaps: # GMT_drywet, GMT_gebco, GMT_globe, GMT_haxby GMT_no_green, GMT_ocean, GMT_polar, # GMT_red2green, GMT_relief, GMT_split, GMT_wysiwyg print "transform to nx x ny regularly spaced native projection grid" # step=5000. step=10000. nx = int((m.xmax-m.xmin)/step)+1; ny = int((m.ymax-m.ymin)/step)+1 topodat = m.transform_scalar(topoin,lons,lats,nx,ny) print "plot topography/bathymetry as shadows" from matplotlib.colors import LightSource ls = LightSource(azdeg = 45, altdeg = 220, hsv_min_val=0.0, hsv_max_val=1.0, hsv_min_sat=0.0, hsv_max_sat=1.0) # convert data to rgb array including shading from light source. # (must specify color m) rgb = ls.shade(topodat, cm.GMT_ocean) im = m.imshow(rgb, alpha=0.15) print "draw coastlines, country boundaries, fill continents" m.drawcoastlines(linewidth=0.25) # draw the edge of the map projection region m.drawmapboundary(fill_color='white') # draw lat/lon grid lines every 30 degrees. m.drawmeridians(np.arange( 0,360,30), color="black" ) m.drawparallels(np.arange(-90,90 ,30), color="black" ) print "draw points" psize=5 font = {'family' : 'serif', 'weight' : 'normal', 'size' : 18} matplotlib.rc('font', **font) x,y = m( lovecraft_lon, lovecraft_lat ) m.scatter(x,y,psize,marker='o', color='white') plt.text(x+50000,y+50000+50000, "Lovecraft", color='white') x,y = m( derleth_lon, derleth_lat ) m.scatter(x,y,psize,marker='o',color='white') plt.text(x+50000-120000,y+50000, "Derleth", color='white', horizontalalignment="right") x,y = m( nemo_lon, nemo_lat ) m.scatter(x,y,psize*3,marker='+',color='#555555') plt.text(x+50000+50000,y+50000-80000, "Nemo", color="#555555", verticalalignment="top") equi(m, nemo_lon, nemo_lat, radius=2688, color="#555555" ) # x,y = m( bransfield_strait_lon, bransfield_strait_lat ) # m.scatter(x,y,psize*3,marker='+',color='#555555') # plt.text(x+50000+20000,y+50000-80000, "bransfield_strait", color="#555555", verticalalignment="baseline") # x,y = m( ross_sea_lon, ross_sea_lat ) # m.scatter(x,y,psize*3,marker='+',color='#555555') # plt.text(x+50000+20000,y+50000-80000, "ross_sea", color="#555555", verticalalignment="baseline") # x,y = m( cape_adare_lon, cape_adare_lat ) # m.scatter(x,y,psize*3,marker='+',color='#555555') # plt.text(x+50000+20000,y+50000-80000, "cape_adare", color="#555555", verticalalignment="baseline") plt.savefig("R'lyeh_locations.png", dpi=600, bbox_inches='tight') # plt.show() 








 French
French Deutsch
Deutsch